Article Text
Abstract
Objectives Air pollution exposure induces cardiovascular effects, possibly via systemic inflammation and coagulation misbalance. Genetic variation may determine individual susceptibility. Our aim was to investigate effect modification by inflammation (Interleukin6 (IL6), tumour necrosis factor-α (TNF-α)) and coagulation (fibrinogen Bβ, plasminogen activator inhibitor-1 (PAI-1)) gene variants on the effect of long-term or short-term air pollution exposure on both blood marker levels and non-fatal myocardial infarction (MI) risk.
Design Population-based case–control study with a nested case-crossover study. Gene-environment interactions for short-term and long-term air pollution on blood marker levels were studied in population controls, for long-term exposure on MI risk using case–control design, and for short-term exposure on MI onset using case-crossover design.
Setting The Stockholm Heart Epidemiology Programme (SHEEP) conducted in 1992–1994 in Stockholm, Sweden. Spatial modelling was used to assess long-term (up to 30 years retrospectively) air pollution exposure to traffic-NO2 and heating-SO2 emissions at home addresses. Urban background NO2, SO2, PM10 and O3 measurements were used to estimate short-term (up to 5 days) air pollution exposure.
Participants 1192 MI cases and 1506 population controls aged 45–70 years.
Outcomes The levels of blood markers of inflammation (IL-6, TNF-α) and coagulation (fibrinogen, PAI-1) and MI risk.
Results We observed gene–environment interaction for several IL6 and TNF SNPs in relation to inflammation blood marker levels. One-year traffic-NO2 exposure was associated with higher IL-6 levels with each additional IL6-174C allele, and 1-year heating-SO2 exposure with higher levels of TNF-α in TNF-308AA homozygotes versus −308G carriers. Short-term air pollution exposure also interacted with IL6 and TNF in relation to marker levels. The risk of MI followed the effect on blood markers in each genotype group.
Conclusions Genetic variants in IL6 and TNF may modify effects of long-term and short-term air pollution exposure on inflammatory marker levels and MI risk.
- Air pollution
- Gene-environment interaction
- inflammation
- IL6
- TNF
This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 3.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/3.0/
Statistics from Altmetric.com
Article summary
Strengths and limitations of this study
-
This is the first study to investigate gene–environment interactions in relation to inflammatory response in a general population, with a simultaneous link to myocardial infarction risk in the same population. We addressed a broad spectrum of air pollution exposure by type (NO2, SO2, PM10, O3) and timing (30 years to 12 h).
-
We primarily focused on identifying consistent interaction patterns, but the multitude of performed tests warrants careful interpretation. Single blood marker measurement was used per participant, and fatal myocardial infarction cases were not included in the study due to the unavailability of blood samples.
Introduction
Cardiovascular disease is a chronic condition associated with inflammation. Both genetic and environmental factors are involved in its pathogenesis. Inflammation is induced by a range of factors including air pollution exposure, and genetic variation may determine the type and intensity of an individual's inflammatory response to environmental stimuli. Air pollutants cause the release of pro-oxidative and proinflammatory mediators from the lungs, and it has been suggested that some constituents, such as the smallest (nanosize) particles, may enter directly into the systemic circulation1 and interact with the endothelium or have direct effects on atherosclerotic plaques and cause local oxidative stress and proinflammatory effects.2
Long-term and short-term exposure to air pollution have been associated with cardiovascular morbidity and mortality.1 We have previously reported in this study population an association between long-term exposure to air pollution and fatal myocardial infarction, especially out-of-hospital deaths.3 The risk of developing MI was also associated with increased levels of blood markers of inflammation (Interleukin6 (IL-6), tumour necrosis factor-α (TNF-α), C reactive protein (CRP)) and coagulation (fibrinogen, plasminogen activator inhibitor-1 (PAI-1)).4–7 Further, in the population controls of our study sample, long-term exposure to traffic-related and heating-related air pollution was associated with increased levels of inflammatory markers (IL-6 and CRP) and short-term exposure to ambient NO2 and PM10 with a suggested increase in IL-6, TNF-α and CRP.8 In another study, investigating MI survivors in six European cities, short-term exposure to particulate matter was associated with increased levels of IL-6 and fibrinogen,9 and certain genetic variants (single nucleotide polymorphisms (SNPs)) in IL6 were also associated with increased IL-6 levels.10 An obvious follow-on question is whether individual genetic make-up can modify an individual's response to air pollution on intermediate biomarkers such as plasma levels of inflammatory and coagulation markers, as well as on the development or acute onset of MI. In MI survivors of the study conducted in six European cities, IL6 (rs2069832, intron) and Fibrinogen Bβ (rs1800790, promoter) variants were indeed seen to modify the effect of air pollution exposure on the levels of IL-6.11 The aim of our study was to investigate, in an adult general population sample and MI cases from the same population, the potential effect modification by genetic variants in genes related to inflammation (IL6, TNF) and coagulation (FGB, PAI-1) on the effects of long-term and short-term exposure to air pollution on blood marker levels and the risk of MI.
Materials and methods
Study population
We studied participants with available data on genotypes and blood marker levels from a population-based case–control study (SHEEP) described in detail elsewhere.12 SHEEP included all first-time MI cases aged 45–70 years identified in the study base (Stockholm County during 1992–1994). Controls matched on sex, age (±5 years) and hospital catchment area were randomly selected from the study base. Gene–environment interaction effects on biomarkers were studied only in the population controls, since the levels of inflammatory and coagulation blood markers in MI cases might be affected by their recent MI and specific medications. Effects on MI for long-term air pollution exposure were assessed using controls and non-fatal MI cases (who survived 28 days and thus provided blood samples for analysis), and for short-term air pollution exposure in MI cases only (table 1). Fatal MI cases and their controls were not included in the study due to unavailability of blood samples. The study was approved by the Ethical Committee at Karolinska Institutet and all subjects gave informed consent.
Overview of the studied populations and statistical methods in relation to exposures and outcomes in the SHEEP study in Stockholm 1992–1994
Risk factors and genotypes
Data on lifestyle factors including heredity, diet, physical activity, job strain, smoking status and other cardiovascular risk factors were collected by questionnaire. Blood samples, blood pressure and anthropometric measurements were obtained during medical examinations at outpatient clinics performed in the morning. Participants were asked not to eat during the preceding night and until the blood sampling. Family history of coronary heart disease (CHD) was defined as having one or more first-degree relatives affected by CHD before age 65. Physical inactivity was defined as reported inactive leisure time, including at most occasional walks, during the last 5–10 years. Participants receiving antihypertensive treatment according to the questionnaire or with systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg at examination were classified as hypertensive. Diabetes mellitus was classified either based on questionnaire-reported use of insulin or other drug or diet treatment, or if newly discovered at examination (reported insulin or drug treatment or elevated fasting blood glucose). Job strain was assessed with the Swedish version of the Karasek-Theorell questionnaire,13 and considered present if a participant scored above 0.765 (75th centile among controls).12 Current smoking was defined as reported smoking within the last year. Participants who had stopped smoking more than 1 year before inclusion were designated as ex-smokers. Routine blood analyses were performed to determine the levels of low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides and insulin. CRP was measured using a high sensitivity immunonephelometric assay (Dade-Behring, Marburg, Germany).4
Selected genes and SNPs were candidate genes for involvement in individual susceptibility to cardiovascular disease, focusing on known SNPs with functional importance. Variants in the genes IL6, TNF, fibrinogen Bβ and PAI-1 have been linked to the increased levels of the respective blood markers,4–6 ,14 ,15 which in turn are associated with increased risk of cardiovascular disease.4–7 At the same time, the inflammatory and coagulation blood markers are involved in a systemic response to air pollution.1 Specifically, we investigated IL6 with SNPs –598G/A (rs180797), –573G/C (rs1800796), –174G/C (rs1800795); TNF with SNPs −1031 T/C (rs1799964), –863C/A (rs1800630), –857C/T (rs1799724), –308G/A (rs1800629), −238G/A (rs3615525); fibrinogen Bβ (FGB) with SNP −455G/A; and PAI-1 with SNP −675 4G/5G. Genetic analyses of IL6 and TNF were performed using dynamic allele-specific hybridisation,4 ,5 and FGB and PAI-1 polymorphisms were analysed with the PCR method.6 ,7
Air pollution exposure
Long-term exposure to source-specific air pollution was estimated at each participant's historical home addresses using a geographical information system as described previously.3 ,16 Briefly, source-specific emission databases were established for 1960, 1970, 1980 and 1990, providing information for dispersion modelling of outdoor levels of locally emitted air pollutants. All addresses inhabited by study participants for more than 2 years were transformed into geographical coordinates, and calibrated dispersion models for air pollution were used to estimate the annual mean level at each address. We calculated average exposures for the preceding 1, 5 and 30 years before study inclusion. Individuals with at least 4 or 25 years of address information were included in the 5–year and 30-year analyses, respectively, and missing air pollution data resulting from unknown residency were replaced by the mean among the controls for that calendar-year.3 For the 1-year mean, missing address information was not accepted. We analysed two air pollutants characterising different exposure sources: NO2 from local traffic emissions (traffic-NO2) as a marker of the mixture of vehicle emissions, and SO2 from local residential combustion heating (heating-SO2) as a marker mainly reflecting oil combustion.
Short-term air pollution exposure measures were based on routine rooftop measurements of urban background NO2, SO2, PM10 and O3 in central Stockholm. Ambient temperature was also measured at the same location. PM10 measurements started in the spring of 1994 and PM10 exposure could thus only be calculated from that point in time. Average exposures to air pollutants as well as ambient temperature were calculated for the intervals of 0–12, 12–24, 0–48 and 0–120 h before the index time point. The index time point was the hour of blood sampling for assessment of effects on blood markers, and the hour of MI symptoms onset or the corresponding control time point for assessment of MI risk in the case-crossover analysis.17 We estimated control intervals for each case using a time-stratified approach: within the calendar month of MI occurrence, all time points on weekdays corresponding to the time and weekday of MI onset were considered as control periods for a case period.18 Exposure intervals with 75% or more of hourly measurements available were included and the interval mean was calculated from the available values.
Health outcomes
Serum IL-6 was measured using an ELISA (IL-6 Eli-pair, Diaclone Research, Besancon, France).4 Serum TNF-α was detected with the Quantikine HS human TNF-α kit (R&D Systems, Minneapolis, New Mexcio, USA).5 An assay of fibrinogen fibrin polymerisation time (FPT test) following a method previously described19 was used to measure plasma fibrinogen levels,6 and the Spectrolyze PAI-1 kit (Biopool AB, Umea, Sweden) was used for determining PAI-1 activity in citrated plasma samples.7
First-time MI cases were identified using standard diagnostic criteria.12 The present study includes only cases who survived 28 days after their MI. The time of MI symptoms onset was determined primarily using information from the medical clinic about the time of symptom onset or admission, or alternatively the admission date was obtained from the National Hospital Discharge Registry. Blood samples were taken from cases and controls about 3 months after disease onset in the case.
Statistical analysis
Linear regression models were used only in the population control sample to estimate effects of air pollution on the levels of blood markers, which were log-transformed to achieve a closer to normal distribution. Estimated air pollution effects were calculated as a change in the log level of an outcome blood parameter (for describing the results, the change in the outcome was back-transformed and presented as per cent change) per standardised unit change in the respective air pollutant.
Logistic regression models were used in the case–control sample, comprising MI cases and population controls, to assess the relative risk of developing MI, estimated as ORs. Effects of short-term air pollution exposure on the risk of MI onset were assessed only in the MI cases using the case-crossover design, with ORs estimated by conditional logistic regression models.
For better comparability across analyses, all air pollution effect estimates are given as per a standardised unit change in the air pollutant, where the unit change corresponds to the difference between the 5th and 95th centiles of the exposure distribution among controls with no missing data (for blood marker analyses) or all intervals with no missing data (for case-crossover MI analysis)—that is, low versus high population exposure.
The analysis of gene–environment interactions was performed in two steps. First, we used likelihood ratio testing (LRT) to assess the significance of models with interaction, compared to models containing just air pollution and genetic variables without the interaction term. Each SNP was assessed in additive (allelic), dominant and recessive genetic models in order to optimise statistical power since these are 1 degree of freedom tests. When a particular SNP was significant in several LRT tests (in studied time intervals and genetic models), we selected the best-fitting model based on the LRT significance level. Second, we performed a graphical assessment to confirm the interaction pattern and best-fitting genetic model based on the data. Thus, figures were created based on an unconstrained genetic model (genotype-specific risk), except for PM10 exposure where, due to lower statistical power, we used the dominant genetic model for the variant allele. In our selected SNPs with significant patterns, the graphical assessment generally confirmed the selected best-fitting models well.
Models describing the genetic interaction with long-term air pollution were adjusted for age and sex in the analysis of blood markers in controls, and for age, sex and hospital catchment area in the case–control analysis of MI (being design variables in SHEEP). Models describing the genetic interaction with short-term air pollution were adjusted for age, sex and ambient temperature except in the case-crossover analysis where we adjusted only for ambient temperature since individual characteristics are controlled for by design. As a sensitivity analysis, significant interaction findings were additionally adjusted for sets of outcome-specific adjustment factors identified previously.3 ,8 Adjustment factors included: age, sex, physical inactivity and level of HDL cholesterol for IL-6 models;8 age and sex for TNF-α models8; age, sex, hospital catchment area, smoking, diabetes, physical inactivity and socioeconomic status for MI models.3 However, the additional adjustment had only a minor impact on the results (data not shown) and did not alter our conclusions. Statistical analysis was performed with STATA V.9 software (StataCorp, College Station, Texas, USA).
Results
Characteristics of the study population are presented in table 2. Family history of CHD, as well as physical inactivity, diabetes mellitus, job strain and current smoking, was more frequent among cases than controls. Levels of IL-6, CRP and PAI-1 were also higher in cases. There were no obvious differences overall in the levels of long-term or short-term air pollution exposure between cases and controls (see online supplementary table S1). However, there was a suggestive non-significant association of short-term exposure to ambient NO2, SO2 and PM10 with slightly increased risk of MI onset (see online supplementary table S2).
Demographics, risk factors and laboratory measurements for non-fatal myocardial infarction cases and controls from the SHEEP study in Stockholm 1992–1994
We investigated two potential pathways for gene–environmental interaction. One of them showed no clear patterns of effect (coagulation FGB and PAI-1 genes), and another had a very clear pattern (inflammation IL6 and TNF genes). Based on LRT tests (data not shown) of effect modification by IL6 and TNF on the association of air pollution exposure with blood marker levels and the risk of MI for the different exposure intervals studied, the strongest and most consistent results were selected, and the patterns of effect across genotypes are illustrated graphically using an unconstrained genetic model (genotype-specific risk) in figures 1 and 2. Estimates for best-fitting genetic models (additive, dominant or recessive) for these associations are exemplified for 1 year average long-term exposure in table 3 and 48 h average short-term exposure in table 4. Similar patterns of interactions were seen for other long-term and short-term exposure intervals (figures 1 and 2), and estimates for best-fitting genetic models for these other long-term and short-term exposure intervals are given in online supplementary tables S3 and S4.
Gene–environment interaction: effect of long-term air pollution exposures on blood marker levels and MI risk among subjects in Stockholm, by IL6 and TNF genotypes
Gene–environment interaction: effect of short-term air pollution exposures on blood marker levels and risk of MI onset among subjects in Stockholm, by IL6 and TNF genotypes
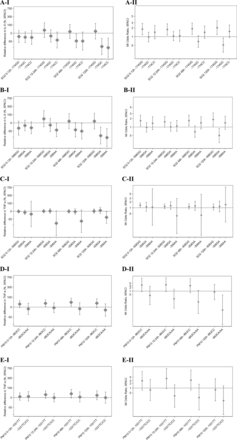
Gene–environment interactions of individual IL6 or TNF SNPs with different measures of long-term exposure to air pollution on the respective blood marker levels in population controls (IL-6, TNF; left panels, ‘I’) and MI risk in the case–control sample (right panels, ‘II’). Effect estimates and 95% CIs for the air pollution effect, by genotype. (A) IL6–174G/C and traffic-NO2. (B) IL6-598G/A and traffic-NO2. (C) TNF-308G/A and traffic-NO2. (D) TNF-308G/A and heating-SO2. Estimates for blood markers (A–D, I) are based on unconstrained (genotype specific) genetic models adjusted for age and sex. Estimates for the risk of MI (A–D, II) are based on unconstrained (genotype-specific) genetic models adjusted for age, sex and hospital catchment area. Effect estimates per 1 unit of change in air pollution exposure, where a unit corresponds to the difference between the 5th and 95th centiles of the exposure distribution in the population controls with no missing data; 28.1 µg/m3 for NO2 1 year, 28.6 µg/m3 for NO2 5 years, 28.8 µg/m3 for NO2 30 years, 6.8 µg/m3 for SO2 1 year, 9.6 µg/m3 for SO2 5 years, 39.4 µg/m3 for SO2 30 years.
Gene–environment interactions of individual IL6 or TNF SNPs with different measures of short-term exposure to air pollution on the respective blood marker levels in population controls (IL-6, TNF; left panels, ‘I’) and risk of MI onset in the cases estimated by case-crossover analysis (right panels, ‘II’). Effect estimates and 95% CIs for the air pollution effect, by genotype. (A) IL6-174G/C and ambient SO2. (B) IL6-598G/A and ambient SO2. (C) TNF-308G/A and ambient SO2. (D) TNF-863C/A and ambient PM10. (E) TNF-1031 T/C and ambient PM10. Estimates for blood markers are based on unconstrained (genotype-specific) (A–C, I) and dominant (D–E, I) genetic models adjusted for age, sex and ambient temperature. Effect estimates per 1 unit of change in air pollution exposure, where a unit corresponds to the difference between the 5th and 95th centiles of the exposure distribution in the population controls with no missing data; 11.6 µg/m3 for SO2 0–12 h, 13.9 µg/m3 for SO2 12–24 h, 11.3 µg/m3 for SO2 48 h, 10.0 µg/m3 for SO2 120 h, 31.8 µg/m3 for PM10 0–12 h, 37.6 µg/m3 for PM10 12–24 h, 34.5 µg/m3 for PM10 48 h, 30.2 µg/m3 for PM10 120 h. Estimates for the risk of MI onset are based on unconstrained (genotype-specific) (A–C, II) and dominant (D–E, II) genetic models adjusted for ambient temperature. Effect estimates per 1 unit of change in air pollution exposure, where a unit corresponds to the difference between the 5th and 95th centiles of the exposure distribution in all intervals with no missing data; 14.0 µg/m3 for SO2 0–12 h, 13.6 µg/m3 for SO2 12–24 h, 11.7 µg/m3 for SO2 48 h, 10.5 µg/m3 for SO2 120 h, 30.9 µg/m3 for PM10 0–12 h, 32.9 µg/m3 for PM10 12–24 h, 26.7 µg/m3 for PM10 48 h, 24.0 µg/m3 for PM10 120 h.
Long-term air pollution showed interactions with IL6 and TNF polymorphisms in relation to the levels of the respective inflammatory blood markers in the population controls and a consistent tendency for the same pattern for MI risk estimated in the case–control sample. For example, long-term exposure to traffic-NO2 interacted with IL6-174G/C in an additive pattern (figure 1A-I), where each additional variant C allele was associated with incremental increase in the air pollution effect on IL-6 levels (table 3). The MI risk analogously increased with each additional variant C allele for all studied exposure windows (figure 1A-II). A similar interaction pattern was seen for the IL6-598G/A SNP (figure 1B, table 3), which was in high linkage disequilibrium (LD) with IL6-174G/C (r2=0.93). For TNF, interaction with long-term air pollution was seen for TNF-308G/A (table 3), with the rare homozygote-308AA being associated with elevated TNF-α levels after long-term exposure to traffic-NO2 and heating-SO2 (recessive genetic pattern) and a consistent pattern seen for MI risk, although the statistical power of this analysis was limited (figure 1C,D, table 3).
The analysis of short-term air pollution exposure interacting with IL6-174G/C and IL6-598G/A resulted in a reverse pattern, where the variant C or A allele was associated with decreased IL-6 levels related to air pollution in the population controls (figure 2A-I and B-I). The corresponding pattern of MI risk across genotypes estimated in cases with a case-crossover analysis was relatively concordant, reflecting higher MI risks from air pollution among subjects lacking the variant allele (common −174GG and -598GG homozygotes; figure 2A-II,B-II). For these interactions, the best-fitting of the investigated genetic models was the dominant model, especially for 2-day and 5-day intervals (see online supplementary table S4). Furthermore, the TNF-308AA genotype was associated with lower levels of TNF-α in population controls after short-term exposure to ambient SO2 (figure 2C-I, table 4); again, the pattern for MI was similar in the case-crossover analysis (figure 2C-II, table 4). Two other TNF SNPs, -863C/A and -1031 T/C (in moderate LD, r2=0.69), showed interactions with short-term PM10 exposure, with common homozygotes having a positive association of air pollution with TNF-α levels while variant allele carriers did not (table 4). This pattern was similar for MI onset (figure 2D-II,E-II).
Discussion
Our findings suggest that systemic inflammatory responses to long-term and short-term air pollution can be modified by genetic polymorphisms in the inflammatory genes IL6 and TNF. Although there was less strong statistical evidence of such interactions in relation to MI risk in the analysis of non-fatal MI cases, MI risk followed the same pattern of effect as for the blood markers. We did not find clear patterns of interaction with air pollution for coagulation genes FGB and PAI-1. Since no blood was available for fatal MI cases, we could not study the link between air pollution, genes and blood marker levels for fatal MI, where long-term air pollution exposure has been shown to play a greater role.3
The main effects of air pollution on blood markers and MI in the studied population were published previously.3 ,8 Briefly, long-term exposure to air pollution was associated with elevated levels of IL-6, while for short-term air pollution exposure a positive, non-significant association was reported for inflammatory markers IL-6 and TNF-α.8 Long-term exposure to air pollution was associated with out-of-hospital death from fatal MI.3 In the present analysis, no significant associations were found for short-term air pollution and non-fatal MI, although point estimates suggested a possible positive effect for all pollutants except ozone (see online supplement table S2).
Our clearest results regarding genetic modification of air pollution effects concern the IL6 gene. Several studies, including our material, have reported an association of elevated IL-6 levels with CHD and MI,4 ,20 and air pollution exposure8 ,9 and genes10 have also been linked to IL-6 levels. However, there is conflicting evidence for an association with MI of the widely studied IL6-174G/C variant.21 In the present study, we observed a gene-environment interaction in a population sample for the IL6-174C allele, with increased IL-6 levels and a consistent trend of increased MI risk after long-term traffic-NO2 exposure. On the other hand, protective effects in terms of lower IL-6 levels and risk of MI onset were noted in carriers of the −174C allele after short-term ambient SO2 exposure. Similar interaction results were seen for IL6-598G/A, in accordance with its high LD with IL6-174G/C. Few studies have investigated IL6 and its interactions with air pollution, with mixed results but suggesting interaction in some form. For instance, IL6 SNP rs2069832 modified the effect of air pollution on IL-6 levels in MI survivors.11 In studies considering smoking, which may be viewed as an inhaled pollutant, the variant −174C allele was associated with increased all-cause mortality risk among 80-year-old participants only in non-smokers,22 while another study reported -174C allele association in middle-aged men with increased MI risk only in smokers.23
In addition, we also saw interactions related to TNF, the other inflammatory gene studied. There have been conflicting reports on the association of the TNF-308G/A polymorphism with MI.24–26 A recent meta-analysis of 24 studies reported an association of the variant TNF-308A allele with developing CHD (OR 1.5, 95% CI 1.23–1.77 for −308A allele carriers compared to common −308GG homozygotes) in Caucasian populations, but not in other ethnic groups.27 Our study showed increased levels of TNF-α in TNF-308AA homozygotes after long-term exposure to traffic-NO2 and heating-SO2, while −308G allele carriers did not show any air pollution effect. However, after short-term exposure to ambient SO2, TNF-308AA homozygotes had slightly decreased TNF-α levels as well as an indication of decreased risk of MI onset. The variant −308A allele has been associated with higher constitutive and inducible TNF-α levels,28 and it is possible that this balance differs after long-term and short-term environmental exposures affecting inflammation and TNF-α levels.
Somewhat surprisingly, we discovered several instances of a reverse pattern of genetic modification by IL6 and TNF SNPs of associations for blood marker levels and MI with long-term versus short-term exposure to air pollution. One potential explanation might be that when particular genotypes contribute to an elevation of inflammatory marker levels over a long period of time, a certain effect of saturation occurs. In this case, additional stimulus may not result in a further increase in the blood marker level. For example, in our previous study of association of IL-6 levels with air pollution exposure, we noted that in current smokers, long-term air pollution exposure does not contribute to an increase in IL-6 levels, compared to never-smokers who respond with elevated IL-6.8 In a healthy organism, IL-6 is expressed in low levels and is controlled by a variety of mechanisms, while its expression is rapidly induced by infection, trauma or other stress conditions.29 During the immediate response to stressors, IL-6 mediates an acute phase response, whereas persistent activity of IL-6 contributes to a switch from acute to chronic inflammation.30 Studies in mice have shown negative effects of prolonged IL-6 exposure. For instance, 1–2 days of IL-6 administration improved hepatic regeneration, while long-term administration of IL-6 (5–7 days) instead sensitised liver cells to injury and death.31 Chronically elevated IL-6 levels also cause hyperinsulinaemia, reduced body weight and liver inflammation in mice.32
In our data, variants in the genes FGB and PAI-1 did not interact with ambient air pollution in relation to the respective markers of coagulation and MI risk. To our knowledge, the PAI-1 gene was not studied previously in terms of such interactions. The FGB variants were investigated in MI survivors and the rs1800790 SNP showed a modifying effect on fibrinogen levels in response to short-term PM10 exposure.33 Additionally, rs1800790 has shown a modifying effect for IL-6 response to short-term exposure to CO.11 Further studies are needed to understand the role of coagulation genes in the cardiovascular effects of air pollution.
The present study helps to elucidate the genetic modification of systemic inflammatory responses to long-term and short-term exposures to air pollution. Such studies have been performed for short-term air pollution exposures in patients with cardiovascular disease,11 but, to our knowledge, our study is the first to investigate such responses in a general population and simultaneously link to MI risk. We had adequate numbers of MI cases and general population controls to investigate gene–environment interactions and to explore causative pathways for air pollution through inflammation to MI risk. Furthermore, we had opportunities to investigate a broad spectrum of air pollution exposures by type and timing. We estimated the health effects (on blood marker levels and MI risk) per the 5th to 95th centile change in the level of the respective air pollutant to facilitate comparability between the different studied pollutants and to quantify effects across a moderate air pollution exposure range. Most of the alternative exposure intervals had a similar range for long-term and short-term exposures, respectively. An exception was long-term heating-SO2 exposure, where a notably higher average and range was seen for 30-year exposure compared to the shorter 5-year and 1-year averages, due to the higher content of sulfur in oil used for heating in the past. An advanced methodology of air pollution estimation by dispersion modelling of emissions, successfully applied previously,3 ,34 ,35 allowed us to study long-term exposures, which are of great importance in the gradual development of atherosclerosis,36 but with a largely unknown relation to inflammation. Assessment of short-term exposure, of potential importance for the acute onset of MI, was based on a well-validated methodology using routine air pollution measurements in the city centre to characterise short-term variation in urban background levels. However, in assessing the effects of individual exposure over hours or days, where commuting time may also be included, inadequate accuracy of rooftop measurements to capture such variation might lead to underestimated associations due to non-differential measurement error. Although currently difficult to undertake on a larger scale, it would be desirable for future studies to incorporate individual measurements using ambulatory equipment in some form.
Our main hypothesis, with a strong biological foundation, relates to the mechanisms of systemic inflammatory response caused by exposure to air pollution.37 Multiple analyses were performed in order to determine consistent interaction patterns and appropriate genetic models, as well as to connect effects on blood markers and on MI risk and onset. We present estimates with CIs and nominal p Values for all analyses. Formal correction for multiple testing was not applied, given the strong mechanistic and biological rationale, and our focus on characterising patterns and mechanisms rather than identifying novel associations. Owing to multiple testing, however, the possibility of chance findings should be considered. We adjusted our analyses for age and sex and, in the case of short-term exposure, also for ambient temperature. As a sensitivity analysis, we assessed outcome-specific sets of further confounding variables,3 ,8 none of which substantially altered our results or conclusions. We had only one single measurement of each blood marker per individual. This is a limiting factor in the interpretation of the results due to the relatively high variability of inflammatory and coagulation marker levels. In future research, repetitive sampling of blood markers could be considered for higher precision. Additionally, both pro-inflammatory and anti-inflammatory markers should be studied, which will give a better understanding of chronic low-grade inflammatory processes induced by air pollution exposure. We were limited to the analyses of non-fatal MI, leaving open the question of whether genetic variants affecting susceptibility to air pollution effects may be differently represented in fatal MI cases. For testing the causative pathway of gene–environment interactions affecting inflammatory marker levels with consequent effect on the risk of MI, it would be advantageous to separately investigate fatal and non-fatal MI as an outcome.
Conclusions
Our results support the concept that genetic variants in IL6 and TNF genes are involved in modifying the effect of long-term and short-term exposure to air pollution on the levels of inflammatory markers in healthy subjects, and that this has consequences for long-term MI risk and MI onset.
Acknowledgments
The authors thank all SHEEP programme participants and the research team, as well as Tomas Lind for his generous help.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Files in this Data Supplement:
- Data supplement 1 - Online supplement
Footnotes
-
Contributors GP and FN initiated and designed the study. UdeF contributed to the design of the initial SHEEP study and was responsible for the clinical investigations and blood samplings. KL was involved in the acquisition of medical data. TB and PL coordinated air pollution exposure assessment. SP analysed the data and drafted the initial manuscript. All coauthors were involved in the interpretation of data, critical revision and approval of the final version of the manuscript.
-
Funding The research was supported by the Swedish Medical Research Council, the Swedish Council for Working Life and Social Research, the Swedish Heart and Lung Foundation, the Swedish Environmental Protection Agency, and King Gustaf the V and Queen Victoria's Foundation.
-
Competing interests FN is employed by AstraZeneca, which also supports his academic part-time work as adjunct Associate Professor.
-
Patient consent Obtained.
-
Ethics approval The Ethical Committee at Karolinska Institutet.
-
Provenance and peer review Not commissioned; externally peer reviewed.
-
Data sharing statement No additional data are available.