Article Text
Abstract
Objectives To evaluate the completeness of reporting of simulation studies on responder analysis methods and simulation performance.
Design Systematic methodological survey.
Data sources We searched Embase, MEDLINE (via Ovid), PubMed and Web of Science Core Collection from inception to 9 October 2023.
Eligibility criteria We included simulation studies comparing responder analysis methods and assessing simulation performance (bias, accuracy, precision or variance, power, type I and II errors and coverage).
Data extraction and synthesis Two independent reviewers extracted data and assessed simulation performance. We used descriptive analyses to summarise reporting quality and simulation performance.
Results We identified seven simulation studies exploring augmented binary methods, distributional methods and model-based methods. No studies reported the starting seed, occurrence of failures during simulations, the random number generator used and the number of simulations. No studies reported simulation accuracy. Responder analysis results were not significantly influenced by covariate adjustment. Distributional methods remained adaptable even with skewed data. Compared with standard binary methods, augmented binary methods generated increased power and precision. When the threshold is in the tail of the distribution, a simple asymptotic Bayesian (SAB) distributional approach may not reduce uncertainty but can improve precision.
Conclusion Simulation studies comparing responder analysis methods exhibit suboptimal reporting quality. Compared with standard binary methods, augmented binary methods, distributional methods and model-based methods may be better choices, but there is no best one.
- Health Surveys
- Review
- STATISTICS & RESEARCH METHODS
Data availability statement
Data are available upon reasonable request.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
This is the first systematic methodological survey to evaluate the completeness of reporting of simulation studies on responder analysis methods and simulation performance.
We give a reference for future researchers if they want to choose one responder analysis method with better statistical performance.
We use a rigorous review process, including comprehensive and systematic search for general medical databases and databases on statistical articles, and two reviewers independently screen, extract data and evaluate all performance measures of each responder analysis method.
Due to different criteria, assumptions and parameters employed in included studies, the variability in simulation approaches across studies limits the inference of our results.
Limited by the simulation performance scale, we cannot give an overall rank of all reported responder analysis methods, leaving the choice of the optimal method unclear.
Introduction
In clinical trials and meta-analysis, the conversion of continuous data into dichotomous data always involves the establishment of a specific threshold.1 This approach, known as responder analysis, categorises participants into ‘responders’ or ‘non-responders’ based on whether their scores surpass or fall below the predefined threshold.2 Once this transformation occurs, the continuous variable is treated as a dichotomous one. The responder analysis aids the interpretation of results and has been widely used in regulatory settings.3 However, responder analysis leads to loss of information and thus reduces statistical power.4
To overcome the potential disadvantages of conventional binary responder analysis methods, other methods have been reported, such as augment binary methods (defining the thresholds considering overall composite end points),5 6 distributional methods (considering responder rate as a function of the mean and SD of the distribution)7 or model-based methods (estimating the responder rate by substituting maximum likelihood estimators of model parameters).8 Other studies proposed responder analysis methods based on data distribution types, such as skew-normal distribution9 or covariables adjustment.10
Simulation studies evaluate the performance of statistical methods by subjecting them to rigorous computer-based procedures in comparison to established truths.11 Simulation studies are increasingly being used in medical research.12 Compared with using alternative statistical methods on observed data, simulation studies evaluate their performance against established truths, offering more compelling evidence regarding the comparative advantages of the investigated methods.13 14 Simulations have been used to compare the performance of responder analysis methods.9 10 15–19 However, no studies have systematically reviewed the reporting completeness and simulation performance of the studies addressing different responder analysis methods. We therefore undertake an evaluation of both reporting and results of simulation studies on responder analysis methods.
Objective
The objectives of this article are to:
Evaluate the reporting quality of simulation studies on responder analysis methods.
Summarise the findings of simulation studies that compare at least two responder analysis methods.
Methods
Search strategy
We systematically searched Embase (via Ovid), MEDLINE (via Ovid), PubMed and Web of Science Core Collection from inception to 9 October 2023, using ‘Responder analysis’, ‘Dichotomous approach’ or ‘Minimal clinically important difference’. We presented the detailed search strategy for each database in online supplemental table S1.
Supplemental material
Study selection
Two reviewers (XC, JR) underwent a pilot screening phase to ensure consistency before formal screening began. They independently screened a subset of studies, resolved discrepancies through discussion and refined the inclusion/exclusion criteria as needed. After this calibration process, they screened titles and abstracts, then full texts, using Covidence (Veritas Health Innovation, Melbourne, Australia). We resolved disagreements through discussion, and if necessary, in discussion with a third reviewer (TL).
We included studies that (1) compared at least two responder analysis methods in at least one simulation and (2) included assessment of at least one of the following properties: bias, accuracy, precision or variance, power, type I and II errors and coverage. We excluded the following types of studies: (1) methodological studies reporting on the development of dichotomisation thresholds, such as the minimal important difference, (2) methodological studies summarising how to calculate dichotomisation, (3) simulation studies that did not report on the simulation performance of responder analysis methods, (4) meeting abstracts, letters, commentaries, editorials, protocols, books or pamphlets and (5) duplicate publications.
Data extraction
Two reviewers (XC, JR) independently abstracted data and resolved disagreements through discussion or, if necessary, with assistance from another statistician (TL). We used an Excel spreadsheet to abstract the following information: (1) general study characteristics, including authors, publication year, country and responder analysis methods, (2) trials information, including medical area of the trial, study design, sample size, outcomes of interest and their definition of thresholds and (3) simulation information: sample size, dichotomisation thresholds and the scenario setting for dichotomisations. Additionally, we noted the author’s conclusions on performance.
Reporting quality assessments
We used the criteria from Zhang et al12 to assess the reporting quality of simulations studies comparing different responder analysis methods.
The defined aims of the simulation.
Simulation procedures:
Reported dependence of simulated data sets.
Reported starting seeds.
Reported random number generator.
Reported the occurrence of failures.
Reported software used to perform simulation.
Reported software to perform analysis.
Justification of data generation.
Investigated scenarios.
Statistical methods.
Number of simulations performed.
Justification for number of simulations.
Criteria to evaluate the performance of statistical methods under different scenarios.
Data analysis
For all analyses, we summarised the categorical variables with numbers and percentages. In addition, we did descriptive analysis for the reporting completeness and simulation performance of reported responder analysis methods.
Patient and public involvement
Patients and the public were not involved in this study.
Results
Characteristics of included studies
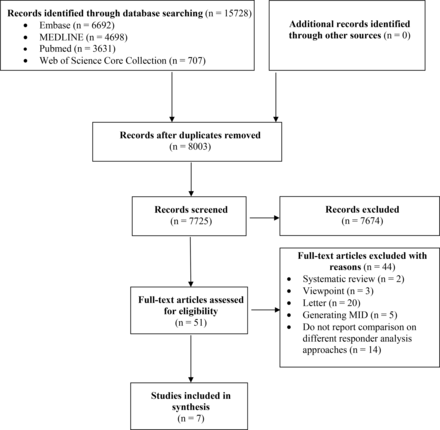
The systematic search identified 15 728 unique records, from which 51 potentially relevant full texts were identified, of which seven simulation studies met the eligibility criteria (figure 1). We presented the basic characteristics in online supplemental table S2. Among them, four studies compared standard binary methods with other responder analysis methods, involving augmented binary methods,17 18 distributional methods16 and model-based methods.19 We presented the details of included responder analysis methods in online supplemental table S3. Sauzet et al focused on different data distribution types. Sauzet et al compared responder analysis for data skew-normal distribution and responder analysis with normal distribution assumption.9 Jiang et al compared the advanced non-asymptotic Bayesian (ANB) approach with the simple asymptotic Bayesian (SAB) approach, simple non-asymptotic Bayesian (SNB) method and traditional beta-binomial (TBB) method.15 Garofolo et al focused on the performance of covariate adjustment in responder analysis and compared responder analysis with adjusted covariates and compared with responder analysis without adjusted covariates.10 The detailed explanation of each responder analysis method is presented in online supplemental table S2. Simulations were conducted based on data from 13 trials of various medical areas, including rheumatoid arthritis, low birth weight, aerobic exercise for pain, capecitabine for cancer, Parkinson’s disease and hyperglycaemic acute ischaemic stroke.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses flowchart. MID, Minimal Important Difference.
Reporting quality of included studies
All studies provided details on the aims of the simulation (online supplemental table S4). Regarding the simulation procedures, all studies reported whether they created independent simulated datasets for different scenarios (ie, situations with different datasets or different data distributions), justification for data generation, scenarios and statistical methods evaluated and criteria used to evaluate the simulation performance. However, none of the studies reported the starting seed, occurrence of failures during simulations or the random number generator used, or explained the reason for the number of simulations. Only one study did not report the software used to perform simulations and analysis, and the number of simulations conducted. The reported software included R, SAS software, and the number of simulation replicates ranged from 1000 to 20 000.
Simulation performance of included studies
Online supplemental table S5 presents the summary of simulation performance of included studies. The details of performance are in online supplemental table S6. Five studies reported power and coverage, three studies reported bias, precision or variance, but no studies reported accuracy.
Standard binary methods with or without adjusted covariates
Garofolo et al10 compared the performance of binary methods using unadjusted covariates and adjusted covariates based on trichotomised baseline severity categories. The comparison aimed to determine whether the adjusted covariates result in less bias than the unadjusted covariates. The findings indicated that there was no substantial difference in the type I error rates and power between the unadjusted and categorically adjusted binary methods. This suggests that adjusting for baseline severity categories did not significantly improve the performance of responder analysis compared with using unadjusted covariates.
Standard binary methods for different data types
Sauzet et al9 investigated the reliability of the distributional methods in handling skewed data. They simulated various sample sizes ranging from 20 to 500 using normal, lognormal, inverse transformation normal and left-skewed normal distribution. The findings indicated that for datasets that were almost normal, the skew normal methods did not perform well unless the sample size was sufficiently large. However, overall, the distributional methods demonstrated applicability for commonly encountered skewed data, enabling researchers to provide both continuous and dichotomised estimates without sacrificing information or precision.
Standard binary methods versus other responder analysis methods
Four studies compared standard binary methods with augmented binary methods, distributional methods and model-based methods. Overall, the compared responder analysis methods have better simulation performance.
In 2016, Wason and Jenkins17 compared the augmented binary methods with standard binary methods in a study on rheumatoid arthritis. Their findings revealed that augmented binary methods allowed for comparable precision with a smaller sample size, without inflating the type I error rate. Overall, the augmented binary methods exhibited enhancements in precision and power, along with reductions in type I error rates and coverage of estimation across different study scenarios.
In 2013, Wason and Seaman18 employed simulated data to compare augmented binary methods with standard binary methods. The results illustrated that the augmented binary methods enhanced precision and reduced type I error rates. Application of the augmented binary methods to data from a phase II cancer trial demonstrated improved precision in estimating success probability, with reduced coverage of estimation.
The study conducted by Peacock et al16 proposed a distributional method as an alternative to standard binary methods of normal distribution data. They simulated four trials to compare the performance of the distributional methods with dichotomisation in terms of bias, power and coverage of CI. In the first trial, which examined birth weight in smokers and non-smokers, the distributional methods outperformed dichotomisation by requiring a smaller sample size to achieve 80% power and providing a CI that was approximately one-third narrower than that of dichotomisation. In the second trial addressing low birth weight and teenage pregnancy, the third trial addressing low birth weight and urinary tract infection and the fourth trial addressing low birth weight and drug use, distributional methods increased power, reduced bias and coverage of CI width for the estimated difference in proportion.
Zhang et al19 conducted a simulation study using data from a Parkinson’s disease study and compared model-based responder analysis methods with standard binary methods. Their findings indicated that compared with usual approach based on dichotomisation, the model-based methods were unbiased in any finite sample and had the potential to reduce sample size while maintaining the same precision, thereby increasing efficiency. Additionally, the model-based methods were found to be more effective in handling missing data.
Different Bayesian responder analysis methods
In 2016, Jiang et al15 developed an ANB approach and compared it with the SAB approach, SNB approach and TBB approach. Through simulations and analysis of pain trial data, they observed improved precision with the ANB approach. However, they also noted that the results may not accurately reflect the true uncertainty, particularly when the threshold was in the tails of the distribution. This highlighted the need for careful consideration of methodological assumptions and potential limitations when applying the ANB approach in data analysis.
Discussion
Key findings
We identified seven simulation studies involving standard binary methods, augmented binary methods, distributional methods and model-based methods. The reporting quality of included simulation studies suffered from failure to report the starting seed, the random number generator, failures that occurred during simulation and providing justification for the number of simulations. No studies reported accuracy. Responder analysis results were not significantly influenced by covariate adjustment. In standard binary methods, distributional methods remained adaptable even with skewed data. Compared with standard binary methods, augmented binary methods generated increased power and precision. Distributional methods produced unbiased estimates, improved power and provided smaller coverage compared with standard binary methods. For missing data, the model-based methods were found to be more efficient with a smaller sample size compared with standard binary methods. When the threshold was in the tail of the distribution, a SAB distributional approach might not reduce uncertainty but improve precision among different types of Bayesian distributional approaches.
Implications
Transparent reporting is crucial since it exposes the limitations of research, thereby facilitating critical appraisal of simulation studies. Our review revealed that none of the included studies explicitly reported simulation ‘accuracy’ as a performance metric. This lack of explicit reporting may hinder interpretation for non-statistical audiences. Future work should consider clarifying the relationship between reported performance metrics and the general concept of accuracy to improve transparency and interpretability. Additionally, statisticians and methodologists need to prioritise enhancing the reporting of simulation procedures for different responder analysis methods by adhering to standards for comprehensive reporting. Finally, there is a need for an approach to ranking the best simulation methods in the future.
For researchers who may use responder analysis to interpret clinical data, in real-world clinical trials, the choice of responder analysis methods depends on a balance between statistical efficiency, interpretability and alignment with clinical goals. Standard binary methods remain widely used due to their simplicity and familiarity among clinicians and regulators. Augmented binary methods improve power and precision, especially in trials with small sample sizes. Distributional methods can accommodate different distributions. Model-based methods are useful in studies with multivariable definitions of response or longitudinal outcome profiles, although they require more statistical expertise and careful model checking. Aligning statistical methods with the clinical context is critical. Trials in chronic or relapsing conditions may benefit from model-based or longitudinal approaches that capture patterns over time, while composite endpoints may offer clinically relevant summaries for complex diseases. Future work should encourage not only methodological innovation, but also practical guidance on when and how each responder analysis method should be used to inform decision-making in clinical research.
Strengths and limitations
Strengths of our study include (1) a comprehensive and systematic search across general medical databases, including databases on statistical articles, (2) two reviewers independently screen and extract data, (3) evaluating all performance measures of each responder analysis method and (4) using an established checklist for simulation studies to evaluate reporting quality.
Our study has several limitations. (1) We do not evaluate the design and conduct of included simulation studies, limiting the quality assessment of included simulation studies. (2) Due to different criteria, assumptions and parameters employed in included studies, the variability in simulation approaches across studies limits the inference of our results. (3) There is no overall rank of all reported responder analysis methods, leaving the choice of the optimal method unclear.
Conclusions
Overall, simulation studies comparing responder analysis methods exhibited suboptimal reporting quality. Compared with standard binary methods, augmented binary methods, distributional methods and model-based methods may be better choices, but we still cannot find which one is the best.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Ethics approval
Not applicable.
Acknowledgments
We thank McMaster Health Science Librarians for their assistance in retrieving articles and facilitating inter-library loans.
Footnotes
Contributors Conceptualisation: XC, DKC, GHG, TL. Methodology: XC, TL. Data collection and analysis: XC, JR. Writing review and editing: XC, DKC, JR, RBP, KY, GHG, TL. Supervision: XC, KY, GHG, TL. XC is the guarantor.
Funding XC is sponsored by The China Scholarship Council – McMaster University joint funding program scholarship (CSC No.202206180008) and 2024-2025 The Eva Eugenia Lillian Cope Scholarship.
Disclaimer The funders had no role in study design, data collection, data analysis and manuscript draft and review.
Competing interests GHG and TL are members of the BMJ Open Editorial Board.
Patient and public involvement Patients and/or the public were not involved in the design, conduct, reporting or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.