Article Text
Abstract
Objective The lack of epidemiological data and molecular diagnostic services in Malaysia has hampered the setting-up of a comprehensive management plan for patients with myotonic dystrophy type 1 (DM1), leading to delayed diagnosis, treatment and support for patients and families. The aim of this study was to estimate the prevalence of DM1 in the 3 major ethnic groups in Malaysia and evaluate the feasibility of a single tube triplet-primed PCR (TP-PCR) method for diagnosis of DM1 in Malaysia.
Design, setting and participants We used PCR to determine the size of CTG repeats in 377 individuals not known to be affected by DM and 11 DM1 suspected patients, recruited from a tertiary hospital in Kuala Lumpur. TP-PCR was performed on selected samples, followed by Southern blot hybridisation of PCR amplified fragments to confirm and estimate the size of CTG expansion.
Outcome measures The number of individuals not known to be affected by DM with (CTG)>18 was determined according to ethnic group and as a whole population. The χ2 test was performed to compare the distribution of (CTG)>18 with 12 other populations. Additionally, the accuracy of TP-PCR in detecting CTG expansion in 11 patients with DM1 was determined by comparing the results with that from Southern blot hybridisation.
Results Of the 754 chromosomes studied, (CTG)>18 frequency of 3.60%, 1.57% and 4.00% in the Malay, Chinese and Indian subpopulations, respectively, was detected, showing similarities to data from Thai, Taiwanese and Kuwaiti populations. We also successfully detected CTG expansions in 9 patients using the TP-PCR method followed by the estimation of CTG expansion size via Southern blot hybridisation.
Conclusions The results show a low DM1 prevalence in Malaysia with the possibility of underdiagnosis and demonstrates the feasibility of using a clinical and TP-PCR-based approach for rapid and cost-effective DM1 diagnosis in developing countries.
- CTG repeats
- genetic counselling
- myotonic dystrophy type 1
- molecular diagnosis
- TP-PCR
- prevalence
This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Statistics from Altmetric.com
Strengths and limitations of this study
This is the first myotonic dystrophy type 1 (DM1) epidemiological study on individuals not known to be affected by DM from the three major ethnic groups in Malaysia.
To date, molecular diagnostic testing for DM1 is not performed in any hospital in Malaysia. This study describes the feasibility of a cost and time-effective triplet-primed PCR-based method for rapid screening and diagnosis of DM1.
The genotyping data does not give allele size accurate to one trinucleotide repeat, but rather is a close approximation of the allele size.
The number of DM1 samples analysed is small as DM1 is a rare disease in Malaysia.
Introduction
Myotonic dystrophies (DM) are the most prevalent adult muscular dystrophy worldwide, with an estimated prevalence of 1 in 8000.1 They are classified into two main subgroups, DM type 1 (DM1) and type 2 (DM2). These are caused by nucleotide repeat expansions, which are inherited as an autosomal dominant trait, and manifest as clinically heterogeneous diseases. DM1 is due to CTG nucleotide repeats beyond the normal length of 5–49, in the 3′ untranslated region of the dystrophia myotonica protein kinase (DMPK) gene, located on chromosome 19q 13.3.2 ,3 It is a progressive disease and categorised into several subtypes. The congenital form of DM1 is maternally transmitted more frequently, although the disease occurs equally in men and women.1 ,4 The general consensus is that the larger the CTG repeat in an individual, the more severe the disease and the earlier the age of onset. It is however, difficult to classify individual DM1 cases into distinct categories based merely on the size of CTG repeats, as genotype–phenotype correlation often overlap and are not clearly defined. In addition, the repeat sizes have shown variation, both between tissues, and over time in the same tissue.5 ,6 This has made disease prognosis difficult. The genetic phenomenon of anticipation can also be observed in the inheritance of the disease, resulting in a more severe form of the disease coupled with an earlier age of onset in subsequent generations.7 ,8
The prevalence of DM1 varies greatly across populations—it is predominantly seen among the Europeans and Japanese.9 ,10 A study also estimated a high disease frequency in the Finnish population.11 In Quebec, Canada, a particularly high DM1 prevalence of 1 in 500 has been recorded due to founder effects.12 In contrast, it is a rare disease among ethnic sub-Saharan populations,13 being almost unheard of with the exception of one case reported in Nigeria.14 Two recent cases among African-Americans have also been observed, most likely representing recent population admixture.15 In view of this disparity, a study was undertaken to determine the distribution of CTG repeats in African individuals not known to be affected by DM1. It was found that there was a highly significant difference in the distribution of normal CTG alleles larger than 18 between the African population and the European and Japanese populations.13 This reiterates a previous theory that CTG alleles between 19 and 30 act as a source of DM1 mutations in subsequent generations.16 These findings have formed the basis for the estimation of DM1 incidence within a population.13 ,17–27
Prior to the establishment of molecular diagnostic tests, DM1 was diagnosed in clinics mainly by observing clinical symptoms and conducting electromyography (EMG) tests, with confirmation by muscle biopsy.28 At present, there are several molecular techniques that can be used in making a DM1 diagnosis, rendering little use for the invasive and painful EMG test and muscle biopsy.29 However, a single test that is able to detect all ranges of expansion sizes is yet to be established. Laboratories often employ a combination of methods depending on mutation dynamics in the population and available equipment. Conventional PCR can detect the normal range of CTG repeats as well as premutated alleles. Optimised PCR conditions can detect alleles up to (CTG)85, whereas those beyond that rely on Southern blot for detection. The triplet-primed PCR (TP-PCR) method was developed to detect the presence of large expanded alleles, thus reducing the number of reflex Southern blot tests.30
As a Southeast Asian country, Malaysia has a population consisting mainly of ethnic Malay, Chinese and Indian. There is also a large group of indigenous people belonging to various tribes. While DM1 has not been frequently diagnosed in this country, there is a possibility of underdiagnosis or misdiagnosis due to the lack of awareness about this condition with its diverse presentations. No study has been performed on the prevalence and incidence of the disease in the predominant ethnic groups, and to the best of our knowledge, diagnostic tests for this disease at the molecular level is not available anywhere in the country. Given the multisystemic and variable phenotypic manifestations in patients, it is therefore important for a simple standard confirmatory diagnostic test to be available, especially when trying to rule out different diagnoses. Here, we report the use of PCR and Southern blot hybridisation methods for the molecular analysis of individuals not known to be affected by DM from the Malay, Chinese and Indian subpopulations, where we studied the length of the CTG alleles in order to predict the prevalence of DM1 in these subpopulations. We also describe the use of a single-tube TP-PCR method for the screening and confirmation of DM1 among Malaysian patients, with the aim of reducing the number of Southern blot tests that need to be performed.
Materials and methods
Ethics statement
The ethics board required that all human participants recruited in the study were briefed on the nature of the study, and provided with an information sheet describing the study. Participants were also assured that their privacy will be protected, and all personal information provided will be kept confidential. Participation in the study was on a voluntary basis, and had no bearing on the quality of care patients received at the hospital.
Sample collection
Blood samples from 377 randomly selected anonymous blood donors not known to be affected by DM of Malay, Chinese and Indian descent were obtained from the University of Malaya Medical Centre (UMMC) blood bank following oral consent to participate in the study. In addition, 11 patients displaying DM-like symptoms were recruited to this study. Written consent, clinical and familial history were obtained from these patients. The ethnicity of participants was determined to be Malay, Chinese or Indian based on their own admission.
Molecular analysis
Genomic DNA was extracted from the blood samples using the QIAamp DNA Blood Mini kit according to manufacturer's protocol (QIAGEN, Hilden, Germany).
Conventional PCR
Analyses of the samples were carried out according to techniques described by Surh et al.31 PCR was performed in a final volume of 30 µL using the Perkin Elmer GeneAmp PCR system. The forward, 103, 5′—CCA GTT CAC AAA CCG CTC CGA GCG TG—3′ and reverse, 96, 5′—GGT GCG TGG AGG ATG GAA CAC GGA C—3′ primers were used. The PCR conditions were set as follows: initial denaturation at 96°C for 5 min, followed by 25 cycles of denaturation, annealing and extension at 96°C, 62°C and 72°C, respectively, for a period of 1 min for each step. Final extension was performed at 72°C for 7 min. The PCR products were sized by gel electrophoresis on 1.5% agarose gel, at 100 V for 45 min. The separated products were cut out from the gel, purified using the QIAquick gel extraction kit (QIAGEN, Hilden, Germany) and sent to a service laboratory for sequencing to determine the exact number of CTG repeats.
Triplet-primed PCR
Analysis of the samples were performed according to techniques described by Singh et al.32 Thirteen samples were subjected to TP-PCR analysis—11 individuals with DM1 symptoms and 2 controls not known to be affected by DM. The participants recruited were all adults between the ages of 30 and 60, and one child aged 5. Testing was performed with 100 ng of genomic DNA from blood samples in a reaction volume of 25 μL. The primers FAM-P1-forward 5′FAM—GGG GCT CGA AGG GTC CTT GT—3′ and P2-reverse 5′—GTG CGT GGA GGA TG AAC ACG—3′ flanked the CTG repeat region, with the forward primer labelled with FAM (fluorescein) fluorescence. The third primer P3 5′—AGC GGA TAA CAA TTT CAC ACA GGA—3′ was designed to bind to the complement of the tail of the fourth primer P4-(CAG)6–reverse 5′—AGC GGA TAA CAA TTT CAC ACA GGA CAG CAG CAG CAG CAG CAG—3′. The primer combination was prepared in a ratio of FAM-P1-forward: P4-(CAG)6–reverse:P3:P2=1.5:1:1.5:1.5, with a final working concentration of 0.6:0.4:0.6:0.6 µM. The TP-PCR conditions were set as follows: initial denaturation at 95°C for 5 min, followed by 10 cycles each of denaturation (97°C) for 35 s, annealing (65°C) for 35 s and extension (68°C) for 4 min. Subsequently, 20 cycles of denaturation, annealing and extension were performed, with the extension time increased by 20 s per cycle to allow for increased yield of PCR product. The products were separated on an ABI PRISM 3130×1 genetic analyser (Life Tech) and fragment size determined using GeneMarker V.2.6 (Softgenetics, State College, Pennsylvania, USA).
Southern blot hybridisation of PCR amplified fragments
Southern blot hybridisation of amplified PCR fragments was carried out in samples that only showed single peaks in the electropherograms, which indicated a CTG expansion or homozygosity for a non-expanded allele. The conventional PCR products were transferred overnight from the agarose gel to a positively charged nylon membrane by capillary transfer and fixing of the DNA to the membrane done via the ultraviolet cross-linking method. The membrane was hybridised overnight in a hybridisation buffer with the addition of 20 µL alkaline phosphatase-conjugated (CTG)10 oligonucleotide at 50°C. The membrane was then removed and the excess liquid drained off, prior to being washed using preheated wash buffers. Following hybridisation and washing of the membrane, the CDP-Star Detection Reagent was applied and the development of the signals was subsequently carried out by exposing the blot to an autoradiography film. Identification of DM1-positive samples were done by comparing the size of the bands or smears obtained with DNA molecular weight markers. The size of the expansion was determined as the point of highest band intensity on the autoradiograph. To estimate the size of the expansion, the lengths of the 100 bp or 1 Kb size marker fragments were first converted into logarithmic values. These values were then graphed on the y-axis against their migration distance on the x-axis. Using linear regression, a line of best fit was drawn through these points and an equation describing that line was derived. The unknown fragment's migration distance derived from the most intense region of the expanded allele was placed into the equation for the regression line to determine a log value for the fragment's size. Taking the antilog of this value yielded the unknown fragment's size in base pairs. This value is an estimate of the average repeat number and is dependent on age at sampling.
Statistical analysis
The frequency of each of the allele present in the 754 chromosomes from the individuals not known to be affected by DM was calculated. Statistical analysis was performed by administering the χ2 test with Yates' correction to compare the distribution of normal large repeats, (CTG)>18, with 12 other populations.
Results
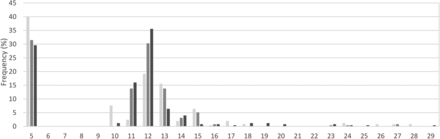
Analysis of DMPK CTG repeat length variation in the general population
The distribution of (CTG)>18 alleles in the Malay, Chinese and Indian subpopulations all point towards a low prevalence of DM1. Figure 1 shows the breakdown of all alleles present in the three subpopulations. (Individual genotyping data is provided in online supplementary files 1–3.) A bimodal allelic distribution was noted—this was in alignment with patterns observed in other populations with low DM1 frequencies. The first peak came from the (CTG)5 alleles, which totalled to 33.7% of all alleles, while the second peak consisted of three alleles, 11–13 that accounted for a majority of 51.1% of the total alleles. The frequencies for (CTG)>18 alleles were 9/250=3.60% (95% CI 0.0166% to 0.0672%) in the Malay subpopulation, 4/254=1.57% (95% CI 0.0043% to 0.0398%) in the Chinese subpopulation, and 10/250=4.00% (95% CI 0.0193% to 0.0723%) among the Indians. Heterozygosity was measured at 79.9%, 77.0% and 76.2% in the three subpopulations, respectively, averaging at 77.7%. This result is aligned to those reported in other populations, which ranged from 73.0% in Europeans17 to 92% in Iranians.26
Frequency of CTG repeats in individuals not known to be affected by DM from the Malay, Chinese and Indian subpopulations. The frequency for large normal alleles, (CTG)>18 was 9/250 or 3.60% in the Malays, 4/254 or 1.57% in the Chinese and 10/250 or 4.00% in the Indians. A bimodal allelic distribution was observed in the Malaysian population, in alignment with patterns observed in other populations with low DM1 frequency. The most frequently seen allele was (CTG)5 in all three subpopulations, whereas (CTG)10–13 was the most common allele group. The genotyping data for each individual is provided in the online supplementary files 1–3. DM1, myotonic dystrophy type 1.
supplementary file
supplementary file
supplementary file
Tables 1 and 2 show the comparison and χ2 analysis of the frequency of (CTG)>18 alleles in individuals not known to be affected by DM1 from the three subpopulations in this study, and in those from 12 worldwide populations, respectively. The (CTG)>18 frequency for the Malay, Chinese and Indian subpopulations were significantly different when compared with frequencies in European, German and Chilean populations. All three Malaysian subpopulations showed frequencies similar to Thai,22 Taiwanese23 and Kuwaiti24 populations. It is also interesting to note that the Han-Chinese show similarity with the Malaysian Chinese, the population that the majority of Malaysian Chinese trace their ancestry to. This allows for our speculation that the DM1 frequency among Chinese Malaysians is low, similar to that observed in the Han-Chinese,24 Taiwanese23 and South African negroids.13
Comparison and χ2 analysis of the frequency of (CTG)>18 alleles in individuals not known to be affected by DM from the Malay, Chinese and Indian subpopulations
Comparison and χ2 analysis of the frequency of (CTG)>18 alleles in individuals not known to be affected by DM from the 3 Malaysian subpopulations to those in 12 worldwide populations
Diagnostic testing for DM1
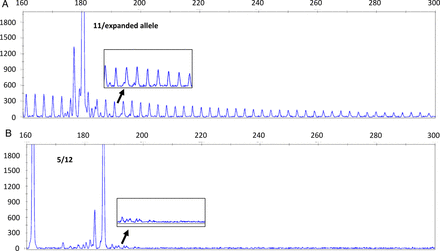
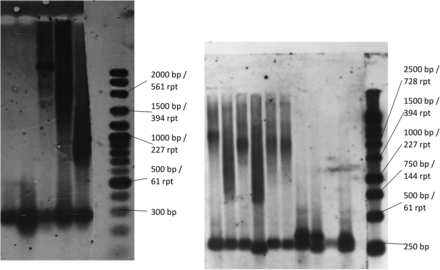
Samples from 11 individuals with DM1-like symptoms and two controls not known to be affected by DM were analysed for CTG expansion using TP-PCR followed by confirmation by Southern blot hybridisation of PCR amplified fragments. TP-PCR testing showed single peaks in nine of the samples, and double peaks in the remaining four. The samples with single peaks also showed a clear laddering pattern indicating the presence of CTG expansion (figure 2). Southern blot hybridisation of PCR amplified fragments confirmed the diagnosis of DM1 in the nine samples, with the detection of expanded alleles ranging from a size of 97–690 CTG repeats, as shown in figure 3. Table 3 shows a summary of the characteristics of the disease exhibited by each patient. Figure 4 shows the pedigree diagram and the CTG repeat size of the families and individuals we studied. It is important to note that apart from those diagnosed (dark squares/circles), none of the other family members were examined or tested for DM1. Hence, there is a possibility that there may be family members showing very mild symptoms who have not presented in our clinics, contributing to the apparent under transmission of the disease in the families.
A summary of the disease characteristics exhibited by patients with DM1 in this study
Electropherogram results of TP-PCR. The x-axis represents the size in base pairs and the y-axis represents the allele peak height. (A) The electropherogram shows a patient sample with DM1 with a single peak corresponding to (CTG)11 and a laddering pattern indicating an expanded allele. (B) Two normal heterozygous alleles with sizes 5 and 12 and no laddering pattern observed. DM1, myotonic dystrophy type 1; TP-PCR, triplet-primed PCR.
Expanded CTG repeats of patients with DM1 Southern blot hybridisation of PCR amplified fragments as seen on an autoradiography film. Expanded alleles in patients ranging from a size of 97 to 690 CTG repeats have been detected. A sample of the bands is shown here, ranging from 270 repeats (1045 bp) to 690 repeats (2305 bp). Normal alleles of four sizes were seen among the patients, 5 (332 bp), 11 (350 bp), 12(353 bp) and 13 (356 bp). Owing to somatic heterogeneity, the expanded alleles usually appear as smears. A 1 Kb DNA ladder as well as samples from individuals not known to be affected by DM were run alongside patient samples as controls. DM, myotonic dystrophy; DM1, DM type 1.
Pedigree diagrams of patients with DM1 studied including the size of their CTG alleles. Members of three families and two individuals had their CTG repeat size analysed. The sizes of the allele pairs for each patient are as stated in the pedigree diagrams. The phenomenon of anticipation was clearly observed in the three families, whereby with the increased CTG expansion in successive generations, a decreasing age of onset is noted. DM1, myotonic dystrophy type 1; Pt, patient.
Discussion
In order to obtain a better understanding of the burden of DM1, we estimated the prevalence of the DM1 using the distribution of CTG alleles larger than 18 in the Malaysian population. The result of (CTG)>18 of 3.05% (23/754) was observed in the Malaysian population. By comparing with the results of studies performed in other populations, we predict that DM1 is a rare disease in Malaysia. A larger study is needed to verify these findings, due to the fact that the participants in this study were recruited from a major hospital in the capital city of Malaysia, therefore may not be representative of the whole country. It is likely that DM1 in the local community is underdiagnosed due to a lack of awareness among the public and healthcare professionals. There are also other contributing factors such as social stigma, and reduced access to major hospitals where specialised consultation and testing are available.
Population studies carried out previously have showed evidence for the association of (CTG)>18 allele frequency and DM1 prevalence. In European populations, the frequency of DM1 is estimated to be 1 in 8000 which corresponded to (CTG)>18 of ∼10%. On the other end of the spectrum, DM1 has only been reported in one Southern African Negroid family where the prevalence of (CTG)>18 is reported to be 0.7%. In the absence of epidemiological data on real cases of DM1, other populations such as the Brazilian, Mexican, Thai, Taiwanese and Han Chinese report the prevalence of DM1 as either higher or lower than populations with known prevalence.
The nine patients identified in this study were diagnosed in the years 2011 and 2012. During these 2 years, the number of patients who were seen at UMMC were 957 418 and 964 497, respectively, totalling to 1 921 915. Using these figures as guide, we estimate the prevalence of DM1 in Malaysia to be <1 in 200 000. This estimate is similar to the low estimates of DM1 prevalence reported in Thai,22Taiwanese23 and Kuwaiti25 populations, where the authors reported the observed frequencies of alleles >18 and correlated them to the prevalence of DM1 in their respective countries. This was also in concordance with the results of our χ2 analysis.
It is interesting to note that the frequency of (CTG)>18 was the lowest in the Chinese subpopulation, although they account for the most number of patients with DM1 seen in our hospital (including those not reported here). Indians on the other hand show the highest frequency of (CTG)>18 in agreement with the findings that DM1 is highly prevalent in India.33 However, the number of Indian patients with DM1 seen in our study was the lowest among the three subpopulations. This may reflect socioeconomic and demographic reasons, as well as misdiagnosis/underdiagnosis of DM1 in the respective subpopulations.
Our study also provides for the first time, data on the (CTG)>18 allele frequency in a Malay population. The Malay ethnic group is genetically more similar to the Chinese compared with the Indians.34 Comparison of the (CTG)>18 distribution of the three ethnic groups, however, shows a closer similarity between the Malays and the Indians (p=0.8151) compared with the Chinese (p=0.249). It would be interesting to see this same analysis performed on other modern Malay populations in the region, such as the Singapore Malays and the Indonesians, as well as the aboriginal Malays.
The usage of the single tube TP-PCR allows for the rapid identification of large pathogenic CTG repeats, thus reducing the need for Southern blot-based approaches to detect or exclude the presence of a large expansion. Southern blot may require large amounts of DNA, the use of radioactive materials and is time consuming. In addition, this procedure is also less sensitive and may be difficult to replicate. Hence, any method that reduces the number of Southern blot that needs to be performed, while demonstrating high sensitivity and specificity is advantageous in a clinical setting. However, the TP-PCR test used requires highly specialised equipment, a genetic analyser, which may not yet be widely available and cannot estimate the size of CTG expansions beyond 85 repeats.
Genotype–phenotype correlation studies in patients with DM1 have thus far given conflicting results, with various underlying mechanisms, associations and theories proposed.35–38 In particular, in their study on the somatic instability of expanded CTG repeats in DM1, Morales et al39 showed that there was no evidence to indicate that pathogenesis of the disease is constrained to a threshold above which repeat length does not contribute towards age at onset. Additionally, they showed that age at onset is further modified by the level of somatic instability, which is a highly heritable trait. In our study, a disparity in the genotype–phenotype correlation in the Chinese family was seen, whereby patient 3 is largely asymptomatic although she carries 350 repeats. Her disease status was only suspected and diagnosed following the birth of her children who exhibited symptoms. Both her children were congenitally affected, which is consistent with findings in previous studies that showed that the majority of congenital cases were maternally transmitted. Patients 2 and 7 on the other hand paternally inherited their pathogenic alleles, resulting in the classic/adult-onset DM1. The same disease phenotype is seen in patients 8 and 9. We were not able to determine whether their diseases were inherited, as their parents have never been tested. However, these patients were given genetic counselling and in accordance with ethical principles, have the autonomy of deciding whether or not to disclose their disease status to family members at risk, for future counselling and testing. It was also observed that congenitally affected patient 5 showed a comparable expansion size to those who were classically affected. The only symptoms he has shown, however, are neonatal hypotonia and a mild cognitive dysfunction. The comparable repeat size is most likely due to the younger age of patient 5 compared with the classically affected adults, and suggests that a larger repeat size would be observed, as the patient grows older. Apart from these disease dynamics, there have also been findings of contraction of allele sizes on transmission reported elsewhere.27 ,36 All these factors point towards the high complexity of DM1 and illustrate the important need for genetic counselling services to be offered to affected families.
Molecular testing is generally established as the gold standard in diagnosing genetic disorders such as DM1. This is because a molecular test is rapidly able to eliminate differential diagnoses, confirm the DM1 diagnosis and estimate the size of CTG expansion in a patient, thus avoiding the need for invasive procedures such as muscle biopsies. Hilbert et al40 who studied a large cohort of patients with DM enrolled in the US National Registry, explored their diagnostic journeys, which on average took 7 years for a correct DM1 diagnosis to be made. This delay brought about many implications to the patients and their families, ranging from lack of appropriate disease management to missed opportunities for genetic counselling. The situation in many developing countries is much similar or even worse as molecular diagnostic testing for DM1 is not easily available. Potentially, there could be a large number of patients who are undiagnosed/misdiagnosed, as well as those who have been unnecessarily subjected to various investigations for a definitive diagnosis to be made.
The findings from our preliminary study can aid the structuring of a rare disease management framework in Malaysia, using DM1 as a disease model. The data presented here add to the scarce literature of DM1 in the Southeast Asian region. The information on CTG repeat lengths of the DMPK gene in individuals not known to be affected by DM, and patents with DM1, together with proper clinical assessment as well as a cost-effective molecular approach, carry implications for earlier diagnosis of DM1 and genetic counselling in a low-resource setting.
Acknowledgments
The authors acknowledge the contributions of Juliana Lee who helped in providing genetic counselling and support to the patients and their families. The authors also thank the UMMC blood bank for blood samples of controls, as well as the patients with DM1 for their participation in this study.
References
Footnotes
KKA, TI and M-KT contributed equally to the manuscript.
Contributors All authors were involved in the conception and design of the work as well as the final approval of the submitted manuscript. KKA and TI were involved in the acquisition and analysis of data and KKA, TI and M-KT were involved in the drafting of the manuscript. LHL, KJG, KTW, AA-A and M-KT contributed to the critical evaluation of the manuscript.
Funding This study was supported by grants from the Ministry of Higher Education, Malaysia (Fundamental Research Grant Scheme; grant number: FG029/2010A), University of Malaya (Postgraduate Research Fund; grant number: PV126/2012A, UM HIR grant number UM.C / 625 / 1 / HIR / MOHE / MED / 27), research grant from the Malaysian Rare Disorders Society and Page Charge Fund from the Institute of Research Management and Monitoring, University of Malaya.
Competing interests None declared.
Ethics approval Ethical approval to conduct this study was obtained from the University of Malaya Medical Centre (UMMC) Ethics Committee (reference numbers 577.17 and 800.6).
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement No additional data are available.