Article Text
Abstract
Objectives To assess the degree to which we can replicate a study between a regional and a national database of electronic health record data in the UK. The original study examined the risk factors associated with hospitalisation following COVID-19 infection in people with diabetes.
Design A replication of a retrospective cohort study.
Setting Observational electronic health record data from primary and secondary care sources in the UK. The original study used data from a large, urbanised region (Greater Manchester Care Record, Greater Manchester, UK—2.8 m patients). This replication study used a national database covering the whole of England, UK (NHS England’s Secure Data Environment service for England, accessed via the BHF Data Science Centre’s CVD-COVID-UK/COVID-IMPACT Consortium—54 m patients).
Participants Individuals with a diagnosis of type 1 diabetes or type 2 diabetes prior to a positive COVID-19 test result. The matched controls (3:1) were individuals who had a positive COVID-19 test result, but who did not have a diagnosis of diabetes on the date of their positive COVID-19 test result. Matching was done on age at COVID-19 diagnosis, sex and approximate date of COVID-19 test.
Primary and secondary outcome measures Hospitalisation within 28 days of a positive COVID-19 test.
Results We found that many of the effect sizes did not show a statistically significant difference, but that some did. Where effect sizes were statistically significant in the regional study, then they remained significant in the national study and the effect size was the same direction and of similar magnitude.
Conclusions There is some evidence that the findings from studies in smaller regional datasets can be extrapolated to a larger, national setting. However, there were some differences, and therefore replication studies remain an essential part of healthcare research.
- Observational Study
- Electronic Health Records
- Retrospective Studies
- DIABETES & ENDOCRINOLOGY
Data availability statement
Data may be obtained from a third party and are not publicly available. The data used in this study are available in NHS England’s SDE service for England, but as restrictions apply they are not publicly available (https://digital.nhs.uk/coronavirus/coronavirus-data-services-updates/trusted-research-environment-service-for-england). The CVD-COVID-UK/COVID-IMPACT programme led by the BHF Data Science Centre (https://bhfdatasciencecentre.org) received approval to access data in NHS England’s SDE service for England from the Independent Group Advising on the Release of Data (IGARD) (https://digital.nhs.uk/about-nhs-digital/corporate-information-and-documents/independent-group-advising-on-the-release-of-data) via an application made in the Data Access Request Service (DARS) Online system (ref. DARS-NIC-381078-Y9C5K) (https://digital.nhs.uk/services/data-access-request-service-dars/dars-products-and-services). The CVD-COVID-UK/COVID-IMPACT Approvals & Oversight Board (https://bhfdatasciencecentre.org/areas/cvd-covid-uk-covid-impact/) subsequently granted approval to this project to access the data within NHS England’s SDE service for England. The de-identified data used in this study were made available to accredited researchers only.
This is an open access article distributed in accordance with the Creative Commons Attribution 4.0 Unported (CC BY 4.0) license, which permits others to copy, redistribute, remix, transform and build upon this work for any purpose, provided the original work is properly cited, a link to the licence is given, and indication of whether changes were made. See: https://creativecommons.org/licenses/by/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
The same team performed the original study and this replication study.
The underlying data sources, while similar, had differences that may have affected the results.
The focus of replication was a single outcome for a single condition and may not generalise to other disease areas.
Introduction
Observational studies using electronic health record (EHR) data are a critical component of the evidence base in population health and epidemiology. However, their findings carry less weight in evidence-based medicine when compared with more conclusive results such as those from randomised control trials. This is partly due to concerns about generalisability and the potential for confounding biases. Replication, the process of repeating a study with a different population or data source, is crucial for strengthening the evidence base in observational research. Successful replication of findings can significantly improve our confidence in their validity and generalisability, leading to a more robust foundation for policy and clinical practice decisions.
Reproducibility is one of the greatest challenges in the area of observational studies.1 2 Goodman et al define three terms for discussing research reproducibility: methods reproducibility, results reproducibility and inferential reproducibility.3 Methods reproducibility is the degree to which a publication includes sufficient information such that other researchers could repeat the analysis. Results reproducibility is the degree to which other researchers can achieve the same results.
We have previously published a study where we compared hospitalisation rates of patients in Greater Manchester (GM) with type 1 diabetes (T1D) or type 2 diabetes (T2D) after contracting COVID-19 when compared with age-matched and sex-matched controls.4 The study reported that following confirmed infection with COVID-19, a number of factors are associated with increased levels of hospitalisation in individuals with T1D and T2D. For patients with T1D, older age, increased social disadvantage, and having hypertension orchronic obstructive pulmonary disease (COPD) were associated with an increased risk of hospitalisation. Other factors were non-significant, potentially due to the small population size. Patients with T2D had the same risk factors as patients with T1D, but with the addition that male sex, non-white ethnicity and severe mental illness had an increased risk of hospitalisation, while taking metformin and low cholesterol levels were associated with a reduced risk of hospitalisation. In this study, we will attempt to replicate these findings in a national database covering the whole of England.
For this replication study, methods reproducibility should have been trivial as we performed the original analysis. However, this was not the case, and in a separate paper, we discuss the methodological factors that make replication problematic, such as differences in the governance, the data structure and the data processing.5 Inferential reproducibility is not possible as it is the degree to which different researchers reach the same conclusions from similar results. Therefore, in this paper, our objective is to assess the degree to which we can achieve results reproducibility between a regional and a national database of electronic health record data in the UK.
If results reproducibility can be achieved, then this will provide evidence that, under certain circumstances, scientific conclusions drawn from regional datasets can be extrapolated nationally.
Methods
Study design
This is a replication of a retrospective cohort study using observational EHR data from primary and secondary care sources in the UK.
Data sources
The data for the original study were from the Greater Manchester Care Record (GMCR). The GMCR is a shared care record containing primary and secondary care data for the residents of GM. The database contains all primary care data, and all hospital admission data, for patients registered to a general practice (GP) in GM who have not opted out of data sharing.
The data for this replication study were from the NHS England National Secure Data Environment (National SDE). The National SDE provides access to a range of national datasets relating to healthcare. Data were made available for COVID-19 research through the CVD-COVID-UK/COVID-IMPACT Consortium which is coordinated by the BHF Data Science Centre and led by Health Data Research UK. The data used for this study were as follows: primary care data from the General Practice Extraction Service Data for Pandemic Planning and Research (GDPPR);6 secondary care data from Hospital Episode Statistics (HES) Admitted Patient Care (APC);7 and COVID-19 test data from the Second-Generation Surveillance System (SGSS) dataset.8 The differences are summarised in table 1.
Differences between the original Greater Manchester study and the two replication studies
Setting
In the original study, all patients from GM (population 2.8 m) with a positive COVID-19 test in their primary care record between 1 January 2020 (month of first UK cases of COVID-19) and 31 May 2021 were eligible.
In this replication study, we have a larger data source. Patients are now from the whole of England (population 54 m after excluding ~1.3 m opt-outs).9 COVID-19 tests are from the SGSS, in addition to those from the primary care record. The date range is now 1 January 2020 to 1 January 2023. The SGSS contains all community COVID-19 test results and so is more complete than the COVID-19 results that appear in a patient’s primary care record.
Approach
We conducted two analyses. Our initial GM study relied on COVID-19 test results that appeared in the primary care record. Therefore, the first analysis was an attempt to reproduce the results of our original study, by only using the COVID-19 test data from the primary care part of the National SDE. The second analysis used the COVID-19 test data from the SGSS, in addition to the primary care data, as this is how researchers using the National SDE would obtain COVID-19 test results.
Study population
For all analyses, the main cohort was defined as patients with a diagnosis of T1D or T2D prior to a positive COVID-19 test result. The controls were patients who had a positive COVID-19 test result, but who did not have a diagnosis of diabetes prior to the date of their positive COVID-19 test result. Each patient in the main cohort was matched with up to three controls. Controls were not reused for multiple patients. Matching was done on age at COVID-19 diagnosis, sex and approximate date (within 2 weeks either side) of COVID-19 test. The date of COVID-19 test is important as outcomes differ depending on the particular wave or variant of COVID-19 that they contracted. Further details of exactly how the cohorts were defined can be found in the original paper,4 and all clinical code lists and analysis code is available here: https://github.com/UoM-Data-Science-Platforms/gm-sde/tree/master/projects/020-Heald.
Variables
The outcome is all-cause hospitalisation within 28 days of a positive COVID-19 test, or in the 2 days prior to account for people admitted to hospital due to COVID-19 but only tested once in hospital. The original study used feeds of admissions data from each hospital within GM. This replication study used the APC table from HES data.
The covariates are a subset of those from the original study. They are the following: year of birth; sex; ethnicity; deprivation via the Townsend score (a measure of social deprivation10); latest values prior to the COVID-19 result for body mass index (BMI), Hba1c, cholesterol (total, LDL and HDL) and eGFR; smoking status; whether the patient has COPD, asthma, a severe mental illness or hypertension; and whether the patient is currently prescribed an ACE inhibitor or ARB, aspirin, clopidogrel or metformin. The covariates in the original study that were not available for this replication study were as follows: latest values prior to COVID-19 result for vitamin D, testosterone and sex hormone-binding globulin. These biomarkers were not available in the GDPPR dataset, which contains a subset of SNOMED concepts from a patient’s primary care record, and therefore were excluded from the analysis. Testosterone and sex hormone-binding globulin had no effect in the original study, while low vitamin D had a small association with increased incidence of hospital admission.
Statistical methods
The original study’s objective was to identify potential factors relating to an increased likelihood of hospital admission in individuals with diabetes, to assess the difference in risk between individuals with and without diabetes and to investigate if any difference in risk could be explained by routinely measured factors. The statistical analysis methods are an exact replication of the previous study.4 A brief overview is as follows.
Modelling was conducted using conditional logistic regression with hospitalisation within 28 days of a positive COVID-19 test as the outcome. We analysed the individuals with diabetes, without the matched controls, using a univariable logistic regression for each factor in turn, for the two groups (T1D and T2D) separately. We then fitted a multivariable model using the patients with diabetes and their controls, with diabetes diagnosis as a covariate and adjusting for sex, ethnicity, Townsend score, hypertension, COPD and BMI.
Following these analyses, we compared the national effect sizes and ORs to our previous work from the GMCR dataset. In addition to a descriptive comparison, we also calculated a conservative 95% CI for the difference between the ORs to find whether there was a statistically significant difference between the effect sizes between GM and the national data.
This analysis was performed according to a prespecified analysis plan published on GitHub, along with the phenotyping and analysis code (https://github.com/BHFDSC/CCU040_01).
Patient and public involvement
The CVD-COVID-UK/COVID-IMPACT Approvals & Oversight Board membership includes five public contributors who ensure that the public/patient voice is considered and embedded appropriately in our projects.
The public contributors review and discuss project proposals (and research outputs) with researchers to ensure work being carried out meets the interests of people affected by heart and circulatory disease or other health conditions, to address any patient and/or public concerns and to advise on best approaches for patient and public involvement throughout the project lifecycle.
Results
Our objective is to demonstrate the degree to which results reproducibility can be achieved. Therefore, all ORs and CIs are displayed visually and discussed descriptively. Full tables with the numeric data for figures 1–4 are available in the supplementary material (online supplemental tables S1–S4).
Supplemental material
Univariable analysis for patients with type 1 diabetes. ‘GMCR’ is the original published study (Greater Manchester Care Record), ‘N1’ is the first replication analysis using COVID-19 test data from the primary care data feed and ‘N2’ is the second replication analysis using the Second-Generation Surveillance System for the COVID-19 test results.
Univariable analysis for patients with type 2 diabetes. ‘GMCR’ is the original published study (Greater Manchester Care Record), ‘N1’ is the first replication analysis using COVID-19 test data from the primary care data feed and ‘N2’ is the second replication analysis using the Second-Generation Surveillance System for the COVID-19 test results.
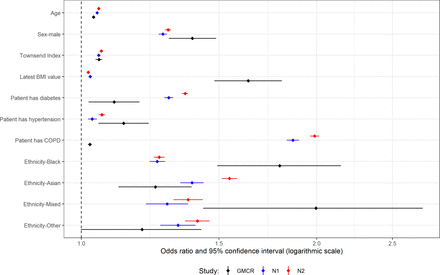
Multivariable analysis for patients with type 1 diabetes and their controls. ‘GMCR’ is the original published study (Greater Manchester Care Record), ‘N1’ is the first replication analysis using COVID-19 test data from the primary care data feed and ‘N2’ is the second replication analysis using the Second-Generation Surveillance System for the COVID-19 test results.
Multivariable analysis for patients with type 2 diabetes and their controls. ‘GMCR’ is the original published study (Greater Manchester Care Record), ‘N1’ is the first replication analysis using COVID-19 test data from the primary care data feed and ‘N2’ is the second replication analysis using the Second-Generation Surveillance System for the COVID-19 test results.
Population comparison
Both national analyses benefited from a much larger population. The original GM study had 862 patients with T1D and a positive COVID-19 test result, while the first national analysis had 38 523, and the second had 77 392 (table 2). The original study had 13 225 patients with T2D and a positive COVID-19 test result, while the first national analysis had 448 829, and the second had 836 532 (table 3). We have previously published a clinical paper focussing on the individuals with T1D.11
Characteristics of the individuals with type 1 diabetes (T1D) and their controls for the three studies
Characteristics of the individuals with type 2 diabetes (T2D) and their controls for the three studies
Most factors analysed were comparable with a few exceptions. Smoking status was much lower nationally (14–15% vs 30–31% for T1D, 12–14% vs 41% for T2D), but this was due to a categorisation error in the original study where anyone with a history of smoking was counted as a smoker. GM is more ethnically diverse, but the GM data also has a higher proportion of unknown ethnicities, possibly because in the National SDE there are more sources of demographic data from which to determine an individuals’ ethnicity. Finally, patients in the national analyses had, on average, shorter lengths of stay in hospital. This is likely due to the later cut-off date for the national analyses, where the combined effect of the reduced severity of later strains and the vaccination programme mean that later diagnoses of COVID-19 are less likely to be severe.
T1D univariable analyses
Out of 25 variables analysed, only three (ACE inhibitor, metformin or mixed ethnicity) showed a statistically significant difference in effect size between GM and the national data (online supplemental table S5). Mixed ethnicity had extremely small numbers in the GM study so the discrepancy here is likely due to random chance and the inconsistencies in reporting mixed ethnicity. For prescribed medications, it is possible that not all metformin or ACE inhibitor SNOMED codes are extracted in the GDPPR dataset which may explain this discrepancy.
All variables that had statistically significant effect sizes in the original study had the same positive or negative association (OR direction) with the outcome in both national analyses (figure 1).
T2D univariable analyses
For the first national analysis, out of 25 variables analysed, only four (latest HDL, COPD, ACE inhibitor, Townsend quintile 2) showed a statistically significant difference in effect size between GM and the national data (online supplemental table S5). For the second national analysis, there were eight that showed a difference (age, cholesterol, eGFR, COPD, ACE inhibitor, clopidogrel, aspirin, Townsend quintile 2).
All variables with statistically significant effect sizes in the original study had the same positive or negative association with the outcome in both national analyses (figure 2).
T1D multivariable analyses
History of COPD and mixed ethnicity were the only variables with a statistically significant difference in effect size between GM and the national data (online supplemental table S6). As mentioned earlier, the original study had very few patients coded as mixed ethnicity and so had a wide CI, and while the ORs do not fall within the original CI, the CIs do overlap (figure 3).
T2D multivariable analyses
For the first national analysis, 8 (out of 11), and for the second, 6 (out of 11) variables showed a statistically significant difference in effect size between GM and the national data (online supplemental table S6).
Most variables have an OR in the national analyses that is outside the CI of the original study (figure 4). However, all ORs are in the same direction as in the original study. Age, Townsend index and hypertension all have a small, but significant, effect size in all three analyses. Being male, or non-white ethnicity, has large effect sizes in all three analyses, though black ethnicity has a smaller OR in the national analyses (first national OR=1.25 and second national OR=1.26 vs GM OR=1.79). Patients with diabetes and patients with COPD have a much larger OR in the national analyses (diabetes: 1.29 and 1.36 vs 1.1, COPD: 1.87 and 1.99 vs 1.03). Latest BMI has much smaller ORs in the national analyses (BMI: 1.03 and 1.02 vs 1.64).
Discussion
We have conducted a study to determine the extent to which results replicate between a regional and a national database of electronic healthcare record data.
EHR data can be variable in quality and contain many unknowns and challenges.12 However, they are typically analysed in large quantities which to some extent mitigates the effects of missingness and noise from random bias. Our analysis has shown that, while the actual ORs from multiple studies may vary, the direction and approximate magnitude of the effect size remain the same. All variables with a statistically significant effect size in the original analysis remained significant, and therefore, clinical decisions made on the results in the regional study would be consistent with the national analyses. This provides some evidence that the findings of regional studies can be extrapolated to a national setting.
However, there were also discrepancies, particularly in the multivariable analysis of patients with T2D and their controls. The large effect size of BMI in the original studies was much lower in the national analyses, and the effect of a patient having diabetes or COPD was much higher in the national analyses. The differences may be due to the underlying data sources, or to differences in the phenotypes as in the GM data the clinical coding was a mixture of Read v2, CTV3 and EMIS codes, while the national database was SNOMED. Therefore, it is important to replicate observational studies in different datasets to better understand the results due to genuine differences between the populations rather than those that are artefacts of the data.
The data analysis code was identical in all studies, but the data curation code was different due to differences in the underlying data. It is therefore possible that differences or mistakes in the data curation code explain some of the discrepancies. All codes used in this analysis are publicly available and therefore open to scrutiny, but it is time consuming for third-party researchers to review this code. In theory, the public nature of the code allows other researchers to identify bugs, but in practice, it is unlikely to occur. One option to discover such errors is for an independent study team to perform the same analysis on the same data. Reproduction of studies using the same data, but performed by a different study team, would be beneficial. However, even that is not a panacea, as discovered in a recent study where 174 independent teams were given the same data and the same research question, yet there was substantial heterogeneity among findings with some showing results with opposite associations with the outcome variable.13
The cohort in the second national analysis was approximately double the cohort for the first national analysis for both T1D (77 392 patients vs 38 523) and T2D (836 532 vs 448 829). The difference between these cohorts was the addition of the SGSS dataset to identify more COVID-19 positive tests. SGSS is a much better source of COVID-19 test data; however, there is no real difference between the results in the two national analyses, suggesting that COVID-19 tests in the primary care record are sufficient for most research.
The original study population appears to have a higher proportion of severe mental illness (SMI) when compared with the national population. The prevalence in GM is likely to be higher than that observed nationally due to the above average levels of deprivation.14 However, in this case, it is predominantly because not all clinical codes used in the original analysis to define SMI were available in the GDPPR dataset and so the apparent prevalence was lower nationally. The original study also had a much higher proportion of smokers. However, this was due to an error where patients who had ever had a current-smoker clinical code in their record were counted as smokers, even if they subsequently had quit. Smoking was therefore excluded from the replication study.
Strengths and limitations
Despite differences in the data sources, the results were remarkably similar, giving strength to the findings in both studies.
The findings in this replication study for this particular disorder may not be transferable to other conditions, although it is likely to be similar for other long-term conditions diagnosed in primary care.
The same researchers conducted both studies and so may have made the same conceptual or procedural errors in both studies.
Knowing the previous study’s results may have subconsciously led us to confirm the previous findings rather than attempt to challenge them.
The replication benefitted from a mix of original researchers and new colleagues from the national SDE, which ensured the replication was as objective as possible.
Conclusion
In two replication studies, performed in a national database, we have shown similar results with a previous study in a smaller, regional database. This provides evidence that results in regional databases can be extrapolated to national settings. However, there were still differences, which further highlights the need for replication of observational studies using electronic health record data, and for different study teams to reproduce work using the same data.
Data availability statement
Data may be obtained from a third party and are not publicly available. The data used in this study are available in NHS England’s SDE service for England, but as restrictions apply they are not publicly available (https://digital.nhs.uk/coronavirus/coronavirus-data-services-updates/trusted-research-environment-service-for-england). The CVD-COVID-UK/COVID-IMPACT programme led by the BHF Data Science Centre (https://bhfdatasciencecentre.org) received approval to access data in NHS England’s SDE service for England from the Independent Group Advising on the Release of Data (IGARD) (https://digital.nhs.uk/about-nhs-digital/corporate-information-and-documents/independent-group-advising-on-the-release-of-data) via an application made in the Data Access Request Service (DARS) Online system (ref. DARS-NIC-381078-Y9C5K) (https://digital.nhs.uk/services/data-access-request-service-dars/dars-products-and-services). The CVD-COVID-UK/COVID-IMPACT Approvals & Oversight Board (https://bhfdatasciencecentre.org/areas/cvd-covid-uk-covid-impact/) subsequently granted approval to this project to access the data within NHS England’s SDE service for England. The de-identified data used in this study were made available to accredited researchers only.
Ethics statements
Patient consent for publication
Ethics approval
The North East - Newcastle and North Tyneside 2 research ethics committee provided ethical approval for the CVD-COVID-UK/COVID-IMPACT research programme (REC No 20/NE/0161) to access, within secure trusted research environments, unconsented, whole-population, de-identified data from electronic health records collected as part of patients’ routine healthcare.
Acknowledgments
This work is carried out with the support of the BHF Data Science Centre led by HDR UK (BHF Grant no. SP/19/3/34678). This study makes use of de-identified data held in NHS England’s SDE for England, and made available via the BHF Data Science Centre’s CVD-COVID-UK/COVID-IMPACT consortium. This work uses data provided by patients and collected by the NHS as part of their care and support. We would also like to acknowledge all data providers who make health relevant data available for research.
Footnotes
X @rwilliams251, @dradrianheald
Collaborators CVD-COVID-UK/COVID-IMPACT Consortium: Jon Boyle, Alastair Proudfoot, Andrew Constantine, Dan Jones, Krishnaraj Rathod, Nida Ahmed, Richard Fitzgerald, Dan O’Connell, Naomi Herz, Rony Arafin, Sonya Babu-Narayan, Zainab Karim, Jon Shelton, Martina Slapkova, Rosie Hinchliffe, Shane Johnson, Renin Toms, Julia Townson, Ewan Birney, Moritz Gerstung, Katherine Brown, Benjamin Zuckerman, Ernest Wong, Tasanee Braithwaite, Anna Stevenson, Annette Jackson, Cathie Sudlow, Fionna Chalmers, Jadene Lewis, James Farrell, Jemma Austin, John Nolan, Kate McAllister, Lars Murdock, Lynn Morrice, Mehrdad Mizani, Melissa Webb, Ross Forsyth, Rouven Priedon, Samaira Khan, Steffen Petersen, Thomas Bolton, Zach Welshman, Caroline Rogers, Alun Davies, Arunashis Sau, Costas Kallis, Fu Siong Ng, Hannah Whittaker, Ioanna Tzoulaki, Jennifer Quint, Juliette Unwin, Libor Pastika, Petter Brodin, Philip Stone, Safa Salim, Sarah Cook, Sarah Onida, Alistair Marsland, Andrew Thompson, Sara Holloway, Thomas Porter, Alastair Denniston, Mamas Mamas, Abdel Douiri, Adejoke Oluyase, Ajay Shah, Alexandru Dregan, Anna Bone, Antonio Cannata, Ben Bray, Charles Wolfe, Daniel Bromage, Dominic Oliver, Elena Nikiphorou, Emeka Chukwusa, Gareth Williams, Gayan Perera, Harry WatsonIrene Higginson, Javiera Leniz Martelli, Jayati Das-Munshi, Joanna Davies, Johnny Downs, Katherine Sleeman, Mevhibe Hocaoglu, Natasha Chilman, Rachel Cripps, Richard Killick, Theresa McDonagh, Vasa Curcin, Carin van Doorn, Rocco Friebel, Arturo de la Cruz, Dorothea Nitsch, Patrick Bidulka, Qiuju Li, Martin Rutter, Adam Hollings, Alistair Jones, Angeliki Antonarou, Daniel Schofield, Deborah Lowe, Elizabeth Kelly, Richardson Richardson, Humaira Hussein, Jake Kasan, Nickie Wareing, Russell Healey, Shoaib Ali Ajaib, Mark Barber, Carole Morris, Felix Greaves, Jennifer Beveridge, Seamus Kent, Thomas Lawrence, Vandana Ayyar-Gupta, Camille Harrison, Myer Glickman, Vahé Nafilyan, Deepti Gurdasani, Frank Kee, Paz Tayal, David Cromwell, Amar Shah, Swapna Mandal, Florian Falter, Joseph Newman, Jennifer Rossdale, Elijah Behr, Nuria Sanchez, Xinkai Wang, Daniel Harris, Amanda Marchant, Ashley Akbari, Daniel King, David Powell, Elizabeth A Ellins, Fatemeh Torabi, Gareth Davies, Hoda Abbasizanjani, Huw Strafford, Jane Lyons, Julian Halcox, Laura North, Marcos del Pozo Banos, Owen Pickrell, Ronan Lyons, Ann John, Robert Aldridge, Abraham Olvera-Barrios, Adnan Tufail, Alasdair Warwick, Alex Handy, Alvina Lai, Ami Banerjee, Ana Torralbo, Ana-Catarina Pinho-Gomes, Andrej Ivanovic, Andrew Lambarth, Anthony Khawaja, Ashkan Dashtban, Becky White, Christina Pagel, Christopher Tomlinson, Chu Siyu, David Selby, Eloise Withnell, Emma Whitfield, Eva Keller, Evaleen Malgapo, Ferran Espuny-Pujol, Flavien Hardy, Floriaan Schmidt, Freya Allery, Harry Hemingway, Honghan Wu, Jinge Wu, Johan Thygesen, Johannes Heyl, Kate Cheema, Katie Harron, Ken Li, Kerrie Stevenson, Laura Pasea, Louise Choo, Luca Grieco, Manuel Gomes, Matt Sydes, Mehrdad Mizani, Michalis Katsoulis, Mohamed Mohamed, Nushrat Khan, Paula Lorgelly, Pedro Machado, Pia Hardelid, Qi Huang, Ravi Shankar, Riyaz Patel, Roy Schwartz, Rui Providencia, Ruth Gilbert, Sam Quill, Samuel Kim, Simon Ellershaw, Sonya Crowe, Spiros Denaxas, Tuankasfee Hama, Waty Lilaonitkul, Yat Yi FanYi Mu, Yoryos Lyratzopoulos, David Osborn, Serban Stoica, Arun Pherwani, Mary Joan Macleod, Sarah Wang, Arun Karthikeyan Suseeladevi, Ben Gibbison, Dann Mitchell, Deborah Lawler, Eleanor Walsh, Elsie Horne, Ewan Walker, Gianni Angelini, Jeremy Chan, John Macleod, Jonathan Sterne, Katharine Looker, Kurt Taylor, Livia Pierotti, Luisa Zuccolo, Martha Elwenspoek, Marwa Al Arab, Massimo Caputo, Mira Hidajat, Neil Davies, Paul Madley-Dowd, Rachel Denholm, Rochelle Knight, Rupert Payne, Shubhra Sinha, Teri-Louise North, Tim Dong, Tom Palmer, Venexia Walker, Yueying Li, Alexia Sampri, Angela Wood, Carmen Petitjean, Chimweta Chilala, Chriselda Oliver, David Brind, Elena Raffetti, Elias Allara, Emanuele Di Angelantonio, Eoin McKinney, Fabian Falck, Genevieve Cezard, Hannah Harrison, Haoting Zhang, Isabel Walter, Jessica Barrett, John Danesh, John Ford, Katie Saunders, Lisa Pennells, Mike Inouye, Robert Fletcher, Rutendo Mapeta, Samantha Ip, Spencer Keene, Stelios Boulitsakis Logothetis, Stephen Kaptoge, Tianxiao Wang, Tom Pape, Wen Shi, Xilin Jiang, Xiyun Jiang, Yanfan Li, Daniel Morales, David Moreno Martos, Huan WangIfy Mordi, Samira Bell, Alan Carson, Alice Hosking, Annemarie Docherty, Athina Spiliopoulou, Baljean Dhillon, Carlos Sanchez Soriano, Caroline Jackson, Christian Schnier, Claire Tochel, Gwenetta Curry, Helen Colhoun, Huayu Zhang, Joe Mellor, Luke Blackbourn, Michelle Williams, Miguel Bernabeu Llinares, Niamh McLennan, Rebecca Reynolds, Richard Chin, Steven Kerr, Tim Wilkinson, Verónica Cabreira, William Whiteley, John Dennis, Angela Henderson, Clea du Toit, Colin Berry, Craig Melville, Deborah Kinnear, Dennis Tran, Filip Sosenko, Frederick Ho, Jill Pell, Jocelyn Friday, John Cleland, Naveed Sattar, Salil Deo, Sandosh Padmanabhan, Terry Quinn, Jianhua Wu, Anna Hansell, Anvesha Singh, Cameron Razieh, Claire Lawson, Clare Gillies, Francesco ZaccardiIain Squire, Kamlesh Khunti, Matthew Bown, Muhammad Rashid, Sharmin Shabnam, Shirley Sze, Tom Yates, Yogini Chudasama, Andrew Mason, Benedict Michael, Caroline Dale, David Hughes, Maria Sudell, Mark Green, Munir Pirmohamed, Reecha Sofat, Rohan Takhar, Ruwanthi Kolamunnage-Dona, Stephen McKeever, Bernard Keavney, Catriona Harrison, Craig Smith, David Jenkins, Evan Kontopantelis, George Tilston, Glen Martin, Hector Chinoy, Joseph Firth, Lamiece Hassan, Lana Bojanić, Matthew Sperrin, Max Lyon, Maya Buch, Richard Williams, Ruth Norris, Ruth Watkinson, Sarah Steeg, Simon Frain, Simon Williams, Camille Carroll, Charlotte Parbery-Clark, Dexter Canoy, Fiona Pearce, Laila Tata, Stephanie Lax, Aashna Uppal, Akshay Shah, Antonella Delmestri, Antony Palmer, Ben Goldacre, Ben Lacey, Dani Prieto-Alhambra, Eva Morris, George Nicholson, Hayley Evans, James Sheppard, Julia Hippisley-Cox, Kazem Rahimi, Linxin Li, Lucy Wright, Mark Ashworth, Marta Pineda Moncusi, Mohammad Mamouei, Nick Hall, Parag Gajendragadkar, Raph Goldacre, Salma Chaudhry, Sara Khalid, Seb Bacon, Seyed Alireza Hasheminasab, Shishir Rao, Zeinab Bidel Taleshmekaeil, Nathalie Conrad, Marie-Louise Zeissler, Jen-Yu Amy Chang, Norman Briffa, Peter Bath, Simone Croft, Suzanne Mason, Tim Chico, Nazrul Islam, Amanj Kurdi, Kim Kavanagh, Marion Bennie, Tanja Mueller, Harry Wilde, Majel McGranahan, Sebastian Vollmer, Christina van der Feltz-Cornelis, Han-I Wang, Lorna Fraser, Tapiwa Tungamirai, Bilal Mateen.
Contributors RW processed and cleaned the data, performed the analysis and drafted the manuscript, and is the guarantor. DJ designed and performed the analysis and reviewed the manuscript. TB assisted with the data processing and cleaning and reviewed the manuscript. AH provided clinical support and guidance on the analysis and discussion and reviewed the manuscript. MM assisted with the data processing and cleaning and reviewed the manuscript. MS designed and performed the analysis and reviewed the manuscript. NP provided overall direction and guidance on the analysis and discussion and reviewed the manuscript. Members of the wider CVD-COVID-UK/COVID-IMPACT consortium (https://www.hdruk.ac.uk/wp-content/uploads/2021/12/211220-CVD-COVID-UK-COVID-IMPACT-Consortium-Members.pdf) also provided comments on drafts of the protocol and manuscript.
Funding The British Heart Foundation Data Science Centre (grant No SP/19/3/34678, awarded to Health Data Research (HDR) UK) funded co-development (with NHS England) of the Secure Data Environment service for England, provision of linked datasets, data access, user software licences, computational usage, and data management and wrangling support, with additional contributions from the HDR UK Data and Connectivity component of the UK Government Chief Scientific Adviser’s National Core Studies programme to coordinate national COVID-19 priority research. Consortium partner organisations funded the time of contributing data analysts, biostatisticians, epidemiologists, and clinicians. The associated costs of accessing data in NHS England’s Secure Data Environment service for England, for analysts working on this study, were funded by the Data and Connectivity National Core Study, led by Health Data Research UK in partnership with the Office for National Statistics, which is funded by UK Research and Innovation (grant ref: MC_PC_20058). This research was co-funded by the NIHR Manchester Biomedical Research Centre (NIHR203308) and the NIHR Applied Research Collaboration Greater Manchester (NIHR200174). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Competing interests None declared.
Patient and public involvement Patients and/or the public were involved in the design, conduct, reporting or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.