Article Text
Abstract
Objective This study developed and validated a stacked ensemble machine learning model to predict the risk of acute kidney injury in patients with acute pancreatitis complicated by sepsis.
Design A retrospective study based on patient data from public databases.
Participants This study analysed 1295 patients with acute pancreatitis complicated by septicaemia from the US Intensive Care Database.
Methods From the MIMIC database, data of patients with acute pancreatitis and sepsis were obtained to construct machine learning models, which were internally and externally validated. The Boruta algorithm was used to select variables. Then, eight machine learning algorithms were used to construct prediction models for acute kidney injury (AKI) occurrence in intensive care unit (ICU) patients. A new stacked ensemble model was developed using the Stacking ensemble method. Model evaluation was performed using area under the receiver operating characteristic curve (AUC), precision-recall (PR) curve, accuracy, recall and F1 score. The Shapley additive explanation (SHAP) method was used to explain the models.
Main outcome measures AKI in patients with acute pancreatitis complicated by sepsis.
Results The final study included 1295 patients with acute pancreatitis complicated by sepsis, among whom 893 cases (68.9%) developed acute kidney injury. We established eight base models, including Logit, SVM, CatBoost, RF, XGBoost, LightGBM, AdaBoost and MLP, as well as a stacked ensemble model called Multimodel. Among all models, Multimodel had an AUC value of 0.853 (95% CI: 0.792 to 0.896) in the internal validation dataset and 0.802 (95% CI: 0.732 to 0.861) in the external validation dataset. This model demonstrated the best predictive performance in terms of discrimination and clinical application.
Conclusion The stack ensemble model developed by us achieved AUC values of 0.853 and 0.802 in internal and external validation cohorts respectively and also demonstrated excellent performance in other metrics. It serves as a reliable tool for predicting AKI in patients with acute pancreatitis complicated by sepsis.
- Machine Learning
- Artificial Intelligence
- Pancreatic disease
- Adult intensive & critical care
- Acute renal failure
- Retrospective Studies
Data availability statement
Data are available upon reasonable request. Datasets generated and analysed during the current study may be obtained upon reasonable request.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
- Machine Learning
- Artificial Intelligence
- Pancreatic disease
- Adult intensive & critical care
- Acute renal failure
- Retrospective Studies
Strengths and limitations of this study
Stacking ensemble combines the predictions of multiple base models to significantly enhance overall predictive performance and generalisation ability while reducing the risk of overfitting.
The Boruta algorithm exhibits greater robustness and flexibility compared with traditional variable selection methods, effectively handling high-dimensional data and non-linear relationships.
The regularisation techniques and hyperparameter optimisation employed in this study can enhance model performance, reduce overfitting and improve generalisation and stability.
This study employs the Shapley additive explanation (SHAP) method to interpret the predictions of the machine learning model and uses the SHAP force plot tool for visualisation, which facilitates understanding and provides information for clinical recommendations.
The limitations of this study include missing values in the original data, a small external validation cohort and the findings based on the intensive care unit limiting their applicability to general wards.
Introduction
Acute pancreatitis (AP) is a common acute abdomen condition characterised by acute inflammation of the pancreas and surrounding tissues, accompanied by abnormal activation and release of pancreatic enzymes, leading to tissue inflammation and necrosis. AP is a complex condition with varying degrees of severity and is a common cause of hospital admission in countries like the USA.1 About 25% of AP patients may progress to severe acute pancreatitis,2 which may involve systemic inflammatory response syndrome (SIRS) and multiple organ dysfunction syndrome. Sepsis is a life-threatening SIRS caused by dysregulated host response to infection, ultimately leading to septic shock and multiple organ failure.3 AP can lead to sepsis,4 with many patients developing AP-related infections in the later stages and severe cases progressing to sepsis.5 6 Studies have shown that the development of sepsis from AP can worsen the condition and increase the risk of mortality.7
In some patients, acute pancreatitis can lead to acute kidney injury (AKI), possibly due to systemic inflammatory response with increased vascular permeability.8 AKI is a common syndrome in intensive care units, characterised by elevated serum creatinine and decreased urine output.9 Several studies have shown a significant increase in mortality rates among AP patients with AKI.10 11 Sepsis-related AKI is also a common condition among critically ill patients, with high incidence and mortality rates.12–14 Therefore, early identification and risk assessment of AKI in patients with acute pancreatitis complicated by sepsis are clinically significant for preventing patient losses and deaths.
Research has explored the influencing factors of AKI in patients with AP and constructed a predictive model. However, there are issues such as small sample size and insufficient model accuracy.15–17 Predicting AKI in septic patients has also been a hot topic in medical research. Some predictive models based on traditional methods, such as logistic regression and Cox proportional hazards model, have been used to predict the development of AKI in septic patients. Fan et al 18 applied logistic regression to construct a predictive model of AKI in 15 726 septic patients, which showed good predictive accuracy. However, the relationship between variables, including linear or non-linear relationships, involves linear regression’s (LR) default handling of linear relationships between independent and dependent variables, which may overly simplify complex non-linear relationships. In addition, LR is susceptible to multicollinearity among variables, which may reduce the model’s performance.19
In recent years, machine learning (ML) has attracted widespread attention from clinical physicians. Machine learning is a branch of artificial intelligence that involves computer simulations or implementations of human learning behaviours. It enables computers to learn from data and improve performance based on their experiences. Machine learning algorithms continuously train to discover patterns and correlations from large databases, then make optimal decisions and predictions based on the data analysis results. Its applications are extensive and commonly used in various fields of medical research,20 such as disease diagnosis, personalised treatment and patient risk prediction. Machine learning algorithms often outperform traditional LR or Cox regression analyses,21 22 as shown in studies like that of Chiofolo et al,23 who used the random forest algorithm to establish a predictive model for AKI in critically ill patients, achieving good early identification of high-risk patients. Yue et al 19 employed machine learning algorithms to construct seven models for predicting the development of AKI in septic patients, aiming to identify the model with the best predictive performance. Currently, risk prediction models for AKI in acute pancreatitis and sepsis are based on fundamental machine learning algorithms such as logistic regression and random forest.24 However, more powerful algorithms like stacked ensemble machine learning (SIML) have not been extensively explored yet.
We noticed a research gap in predicting AKI in patients with AP combined with sepsis. AP patients often develop sepsis in the intensive care unit (ICU), resulting in higher mortality rates. However, early and accurate diagnosis of AKI in AP patients with sepsis remains challenging. Therefore, this study used a large database to develop and validate a superior performing stacked ensemble machine learning predictive model. The aim was to predict the occurrence of AKI during ICU hospitalisation in patients with AP complicated by sepsis, using key risk factors determined through feature selection. This model can assist clinicians in assessing the risk of acute kidney injury in patients and implementing appropriate interventions and treatment measures, thus, achieving early intervention and treatment goals.
Method
Data source
The research data originates from the MIMIC database, and this study is a retrospective cohort study. The MIMIC database is an open database system based on a large biomedical dataset, primarily used to simulate patient conditions in the ICU. (dataset)MIMIC-III25 collected data from 53 423 adult patients admitted to the ICU at Beth Israel Deaconess Medical Centre (BIDMC) from June 2001 to October 2012, as well as data from 7870 neonatal intensive care patients admitted from 2001 to 2008. The (dataset)MIMIC-IV database,26 an improvement over MIMIC-III, gathered clinical data from over 190 000 patients and 450 000 hospitalisations at BIDMC from 2008 to 2019. The database records detailed information such as patients’ demographic data, laboratory tests, medication records, vital signs, surgical procedures, disease diagnoses, medication management, follow-up survival status and more. We used patient data from the MIMIC-IV database for model development and internal validation, followed by external validation using patient data from the MIMIC-III database. All patient information in the database has undergone de-identification processing, eliminating the need for individual patient consent or ethical review board approval.
Study population
We first extracted data from the MIMIC database for all patients diagnosed with acute pancreatitis. Subsequently, we further screened this data to select patients who met the diagnostic criteria for sepsis, which served as the target population for the subsequent study.
Our study aimed to assess the occurrence of AKI within 7 days of ICU admission in patients with AP combined with sepsis. According to the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3), sepsis patients were screened based on the presence of documented or suspected infection and a Sequential Organ Failure Assessment (SOFA) score greater than 2. The SOFA score (Sequential Organ Failure Assessment score) is a clinical scoring system designed to assess the degree of multiple organ dysfunction in critically ill patients. The SOFA score is based on the functional status of six major organ systems: respiratory, circulatory, hepatic, neurological, renal and haematological systems, with each system assigned a score corresponding to its functional state. Specifically, the scoring ranges from 0 to 4, where 0 indicates no dysfunction and 4 signifies the most severe organ dysfunction. The total SOFA score can be used to evaluate the overall condition of the patient. A total score greater than 2 is generally considered to indicate an increased risk of organ dysfunction, with higher scores correlating with a greater likelihood of poor prognosis.
According to the Kidney Disease: Improving Global Outcomes (KDIGO) classification system for kidney diseases, AKI was diagnosed if there was an increase in serum creatinine (Scr) of more than 0.3 mg/dL within 48 hours, an increase of ≥1.5 times baseline within 7 days or urine output less than 0.5 mL/kg/h for more than 6 hours. The first Scr value on admission was used as the baseline Scr, and AKI was evaluated based on the worst serum creatinine and urine output within 72 hours of suspected sepsis diagnosis and ICU admission. We excluded patients who were under 18 years old, had a SOFA score less than 2, were admitted to the ICU for less than 24 hours, already had AKI on ICU admission or had a history of renal failure. The flowchart is depicted in figure 1.
Research workflow diagram.
Data extraction
This study extracted rich variables from multiple aspects, including (1) demographic information: age, gender, race, weight, height and comorbidities such as cerebrovascular disease, diabetes and chronic lung disease; (2) vital signs: including heart rate, respiratory rate, systolic blood pressure, diastolic blood pressure, partial pressure of carbon dioxide, partial pressure of oxygen and oxygen saturation; and (3) laboratory test indicators (including maximum and minimum values): albumin, anion gap, lymphocytes, neutrophils, monocytes, bicarbonate, bilirubin, serum calcium, serum chloride, serum potassium, serum sodium, serum creatinine, haematocrit, haemoglobin, lactate dehydrogenase, serum magnesium, mean corpuscular haemoglobin concentration, pH value, platelets, prothrombin time, activated partial thromboplastin time, blood urea nitrogen, white blood cells, red blood cells, glucose, lactate, Glasgow Coma Scale score and urine output. Additionally, factors such as whether vasopressors were used, whether continuous renal replacement therapy was received, whether invasive mechanical ventilation was used and whether antibiotics were used were also considered. The comprehensive consideration of these variables provides robust data support for the study, aiding in a more accurate assessment of the risk factors for AKI in patients with AP complicated by sepsis.
To reduce the bias caused by missing data, we used the missingno module in Python 3.10 software to filter out missing data. In figure 2, each column represents a clinical variable, with white lines indicating missing data. The more white lines in a column, the more missing values for that variable. We excluded variables with a missing rate exceeding 30%, such as height and serum albumin levels, to ensure the accuracy of the study and models. The remaining variables’ missing values were imputed using the mice package in Python software for multiple imputation (MI). Multiple imputation is an effective statistical method that allows for reasonable estimation of missing data while preserving the structure and characteristics of the dataset. We performed subsequent analyses based on the multiple imputed datasets generated, thereby, reducing bias associated with missing values and enhancing the robustness and credibility of the results. Vital signs and relevant laboratory parameters were characterised using maximum and minimum values, treated as independent features and included in the study.
Missing data distribution plot. (Each column represents a clinical variable, with white lines indicating missing data. The more white lines in a column, the more missing values for that variable.).
Statistical analysis
This study used R (version 4.3.2) statistical software for data analysis. For continuous variables following a normal distribution, the mean and SD were used for representation, while for non-normally distributed variables, the median and quartiles were used. Categorical variables were represented using percentages. In inter-group comparisons, paired t-tests or Mann-Whitney U tests were employed for continuous variables, while χ² tests or Fisher’s exact tests were used for categorical variables.
The variable selection stage employed the Boruta algorithm. This algorithm operates by randomly sampling the original features and generating random features to construct a random forest model. Using the random forest algorithm, it computes the importance score (z-value) for each original feature and generates a set of ‘shadow’ features combined with the original ones. Subsequently, it calculates the importance score for the combined features and identifies important features by comparing the z-values of the original and ‘shadow’ features. Through recursively removing non-important features and recalculating the importance scores of the new feature sets until all features are classified as important or non-important. Finally, variables with a p value <0.05 are selected as inputs for subsequent analysis.
Results
Baseline characteristics
Patient baseline characteristics differences are shown in table 1. During ICU hospitalisation, male patients were more prone to AKI than female patients. AKI patients had higher age and weight; higher incidence of comorbidities such as diabetes and chronic lung disease; and a greater number of individuals using antibiotics, vasopressors and mechanical ventilation compared with non-AKI patients. The average length of hospital stay for AKI group patients was 13.95 days, with an average ICU stay of 4.59 days, significantly higher than the average hospital stay (7.66 days) and average ICU stay (2.23 days) for non-AKI group patients.
Comparison of patient baseline data
Feature selection
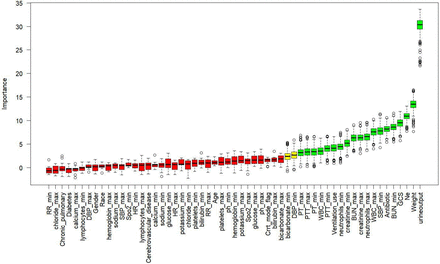
We excluded several variables from the original data, such as alkaline phosphatase, alanine aminotransferase and aspartate aminotransferase, due to the presence of high collinearity issues in the assessment of liver function. The results of feature selection based on the Boruta algorithm are shown in figure 3. We employed the following parameter settings: first, we specified the model formula in the form of ‘status ~ .’ where ‘status’ refers to the outcome event to extract all variables from the data and treat them as independent variables. To ensure significance, we set the p value threshold at 0.05 and enabled multiple comparison adjustments (mcAdj=TRUE) to reduce the type I error rate using the Bonferroni method. Furthermore, we established the maximum number of iterations at 500 (maxRuns=500) to enhance the stability and accuracy of feature selection, while setting the detailed output during runtime to 0 (doTrace=0) to keep the results concise. To preserve the importance history for each iteration, we set the holdHistory parameter to TRUE, allowing us to analyse the variability of variable importance across different iterations. Lastly, to compute feature importance, we used the getImpRfZ function, which provides an effective assessment of feature importance by running a random forest model and collecting the Z scores of average accuracy decreases. The entire process was conducted using RStudio software.
Feature election ased on Boruta lgorithm. (The x-axis represents the names of each variable, while the y-axis represents the z-values of each variable. Green boxes denote important variables selected by the algorithm, yellow boxes represent tentative variables and red boxes indicate unimportant variables. BUN, blood urea nitrogen; DBP, diastolic blood pressure; GCS, Glasgow Coma Scale; HR, heart rate; INR, International Normalised Ratio of Coagulation Function; PT, prothrombin time; PTT, partial thromboplastin time; RRespiratory ate; SBP).sba, rr, systolic blood pressure
The 19 variables most closely associated with AKI are weight, GCS (Glasgow Coma Scale), urine output, vasopressin, mechanical ventilation, antibiotics, minimum systolic blood pressure (SBP), minimum and maximum white blood cell count (WBC), minimum and maximum blood urea nitrogen (BUN), minimum and maximum serum creatinine, minimum and maximum neutrophils count, minimum and maximum prothrombin time (PT) and minimum and maximum partial thromboplastin time (PTT).
Model development
After completing feature selection, the machine learning predictive model was developed using Python (3.10).
The AKI screening data from the MIMIC-IV database was randomly allocated to training and testing datasets in a 7:3 ratio. The training dataset was used for algorithm development, while the testing dataset was employed to evaluate algorithm performance. Initially, eight machine learning models were constructed, including logistic regression, support vector machine, CatBoost, random forest, XGBoost, LightGBM, AdaBoost and MLP (multilayer perceptron). To optimise the overall model performance, we implemented feature selection during the modelling process to reduce model complexity and enhance generalisation capability. Additionally, we introduced some regularisation methods such as L1 and L2 to address overfitting issues. In online supplemental file 1, we describe in detail how to use regularisation techniques when building machine learning models. In online supplemental file 2, we present the parameters used in the construction of each model, along with detailed explanations.
Supplemental material
Supplemental material
Based on these models, a Stacking ensemble model named ‘Multimodel’ was built. Hyperparameter tuning was conducted using GridSearchCV, wherein a parameter space was defined for each model, allowing for training with various parameter combinations to select the optimal hyperparameter set. Stratified K-Fold cross-validation was employed to ensure that the class distribution remained consistent across each training and testing split; all models in this study used 10-fold cross-validation. The models were ultimately trained by inputting the training data along with the parameter space to identify the model with the best performance.
We implemented the model fusion process (Ensemble Learning) using a Stacking Classifier. First, we created a list of model names, ‘model_names’, and used joblib to load the eight previously trained models via the load function. We then defined a collection of base models, ‘estimators’, composed of the loaded models, each associated with its respective name. The base models included logistic regression (Logit), support vector machine (SVM), classification tree (Cat), random forest (RF), XGBoost, LightGBM, AdaBoost and multilayer perceptron (MLP). During the construction of the ensemble model, iterative computations revealed that using logistic regression as the meta-learner yielded the best results, Therefore, logistic regression (LogisticRegression) was used as the final model (final_estimator). By calling the fit method, the training data (x_train) and the corresponding labels (y_train) were input into the stacking classifier (clf). The stacking classifier automatically trains the base models internally and uses their predicted results as new features for the final model to make the ultimate predictions. This process enhances classification performance through the method of model fusion. By leveraging the strengths of multiple base classifiers, the stacking classifier integrates the outputs of these models, thereby, improving the overall accuracy and robustness of the predictions. In this process, labels such as ‘model_names’, ‘estimators’, ‘x_train’ and ‘y_train’ are defined by us and can be modified as needed. Figure 4 illustrates the algorithmic composition of the stacked ensemble model (‘Multimodel’). In online supplemental file 3, we demonstrates the performance of the ensemble model when using different base models as meta-learners.
Supplemental material
Composition of stacked ensemble model algorithm.
Model validation
To validate the generalisation and predictive ability of the stacked ensemble model we constructed on other datasets, we first conducted internal validation using 30% of the testing dataset from the MIMIC-IV database. Subsequently, we treated MIMIC-III as an independent database and used its data for external validation of the model. During the external validation process, we applied the model to patient data from the MIMIC-III database and assessed its performance and generalisation ability. By combining internal and external validation, we were able to comprehensively evaluate the performance of the constructed stacked ensemble model on different datasets, providing reliable support and guidance for further clinical applications.
Performance comparison of the model on the internal validation set
We developed nine machine learning models to predict the development of AKI in patients. Figure 5 displays the discriminative performance of these nine models on the ROC curve. The ROC curve, also known as the receiver operating characteristic curve, is a comprehensive indicator reflecting the sensitivity (true positive rate) and specificity (true negative rate) of continuous variables. It describes the classifier’s performance changes at different thresholds. The closer the curve is to the upper-left corner, the better the classifier’s performance. In practical scenarios, due to sample imbalance, the curve may lean towards a certain class. In such cases, the area under the curve (AUC) is used to evaluate the classifier’s performance. AUC closer to 1 indicates better classifier performance. In addition, we evaluated and compared the model’s performance using metrics such as precision, accuracy, recall and F1 score.
Receiver operating characteristic curves of the nine models.
Among the eight base models, the Logit model (AUC=0.824, precision=0.865, recall=0.749, F1 score=0.803) demonstrated better predictive performance for AKI in AP patients with sepsis, followed by the RF model (AUC=0.822, precision=0.794, recall=0.877, F1 score=0.833), SVM model (AUC=0.821, precision=0.875, recall=0.696, F1 score=0.775), CatBoost model (AUC=0.815, precision=0.851, recall=0.766, F1 score=0.806), XGBoost model (AUC=0.813, precision=0.761, recall=0.930, F1 score=0.837), MLP model (AUC=0.812, precision=0.810, recall=0.871, F1 score=0.839), AdaBoost model (AUC=0.805, precision=0.802, recall=0.830, F1 score=0.816) and LightGBM model (AUC=0.803, precision=0.803, recall=0.813, F1 score=0.808). Using the SVM model (AUC=0.821) as a reference, both the Logit and RF models exhibited superior predictive abilities for AKI in AP patients with sepsis, while the abilities of the CatBoost, XGBoost, MLP, AdaBoost and LightGBM models were inferior to the SVM model. The performance of the ensemble model (Multimodel) surpassed that of any single base learner, with an AUC value as high as 0.853 (0.792–0.896), indicating stronger predictive capability. Table 2 provides detailed performance metrics for the nine models.
Performance metrics of the model on the internal validation set
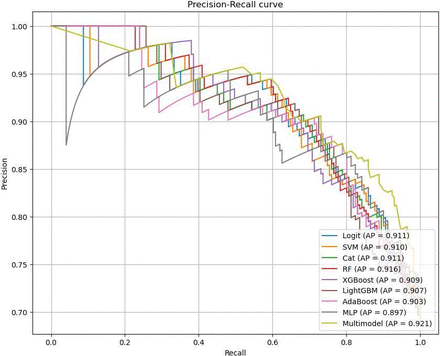
The discriminative performance of the ensemble model (Multimodel) was the best, with the highest accuracy (0.798) and F1 score (0.853). From the precision-recall curve (figure 6), it can be observed that the Multimodel outperformed other models, demonstrating better classification performance and suggesting it as the optimal model with significant clinical utility.
Precision-recall curve. (The relationship between precision and recall is described, where higher values indicate better classification performance of the model.).
Performance comparison of the model on the external validation set
Online supplemental table 1 presents detailed performance metrics for the nine models on the external validation set.
Supplemental material
During the external validation stage, the stacked ensemble model exhibited an AUC value of 0.802 (0.732–0.861), an accuracy of 0.715 and an F1 score of 0.834.
Model Interpretability
We developed a stacked ensemble model named ‘Multimodel’ comprising eight machine learning algorithms. Given the opaque black-box nature of machine learning, we employed the Kernel-SHAP method for model interpretation. The Kernel-SHAP method estimates the influence of features on prediction outcomes by computing the expected marginal contributions of feature values. It uses a kernel function to approximate Shapley values, thereby, avoiding the computational complexity of considering all possible feature subsets.
Through an analysis of feature importance based on Shapley additive explanation (SHAP) values, we demonstrated the 19 predictive factors crucial for AKI occurrence (online supplemental figure 1a) and ranked the feature importance for each variable. Additionally, via a summary plot of SHAP values (online supplemental figure 1b), we described the contributions of each predictive factor to the outcome. In this plot, SHAP values exceeding 0 indicate an increased risk of AKI occurrence, while values below 0 indicate a decreased risk.
Supplemental material
The SHAP force plot visualises the model predictions as the results of feature contributions. By demonstrating how the stacked ensemble model generates predictions for four representative individuals, the model provides clinicians and patients with intuitive guidance, enhancing their understanding of how the model makes specific predictions.
In the internal validation set, we randomly selected four samples for individualised prediction of AKI. Online supplemental figure 2a,b display the SHAP force plots for two patients who experienced AKI. According to our model predictions, the first patient developed AKI during ICU stay, with a weight of 124 kg, vasopressor use during hospitalisation, a urine output of only 1546 mL, a minimum creatinine value of 55 µmol/L and a maximum white blood cell count of 22.1×109 /L, all higher than normal levels. On the other hand, the second patient who experienced AKI had a urine output of 1259 mL, a minimum PTT value of 23.6 s, a maximum creatinine value of 15 µmol/L, a minimum neutrophil count of 10.058×109 /L and a minimum white blood cell count of 9.9×109 /L, all of which increased the risk of AKI occurrence. Online supplemental figure 2c,d depict two patients who did not develop AKI. The first patient had a maximum white blood cell count of 3.8×109 /L, a minimum creatinine value of 7.0 µmol/L and a minimum neutrophil count of 4.3092×109 /L, significantly lower than the levels seen in patients who developed AKI. This patient had a urine output of 2645 mL, higher than that of patients who developed AKI, and a GCS score of 14. According to the model prediction, this patient did not develop AKI. Meanwhile, the second patient who did not develop AKI had a weight of 73 kg, a maximum white blood cell count of 6.8×109 /L, a minimum creatinine value of 6.0 µmol/L, a minimum systolic blood pressure of 111 mm Hg, a urine output of 2200 mL and a GCS score of 14. The actual outcomes were consistent with the model predictions.
Discussion
In identifying high-risk patients for AKI, the application of artificial intelligence and machine learning algorithms is somewhat limited, often built on the foundation of previous-generation models. Compared with more advanced algorithms, their accuracy tends to be relatively limited. To enhance predictive efficiency, this study proposes the use of the SIML method, which involves integrating multiple algorithms.
Integrated technology refers to the utilisation of various machine learning algorithms or models to generate an optimal predictive model. Compared with using individual base learners alone, the generated model typically exhibits better performance. Integrated technology mainly includes methods such as Bagging, Boosting and Stacking. In this study, Stacking method was chosen. Stacking technology can combine the prediction results of various models to form a more powerful meta-learning model. In this approach, the meta-learner uses predictions from different weak learners as feature inputs, learning how to best combine these input predictions to produce superior output predictions. By parallelly integrating predictions from different models, Stacking technology helps improve prediction accuracy, reduce variance, mitigate overfitting and enhance the robustness of the model.27 The process of stacking technology involves two key levels: first, training multiple independent machine learning models in the first level to obtain their respective performance scores; second, composing a meta-learner in the second level that uses the predictions of the first-level models for training to enhance overall performance. Stacking method has the characteristic of integrating multiple classifiers while ensuring excellent performance.
This study is based on the MIMIC database and uses the Boruta algorithm for variable selection. Subsequently, multiple machine learning algorithms are employed to construct a prediction model, and a stacked ensemble model is successfully developed. Through repeated computations and validations, our model demonstrates excellent performance in both the training and validation sets. We constructed a prediction model based on 19 key variables selected by the Boruta algorithm. In internal validation, the AUC value of this model reached 0.853, and it also performed well on other evaluation metrics. In external validation, our model achieved an AUC value of 0.802, indicating good generalisation ability and effective prediction of the risk of AKI occurrence in AP combined with Sepsis patients. These results fully demonstrate the reliability and effectiveness of our model, providing important reference value for further research and clinical practice.
We use SHAP values to reveal the black box of machine learning. SHAP values28 are a technique for explaining the prediction results of machine learning models. By calculating the contribution of each feature to the model’s prediction results, SHAP values reveal the prediction process of the model. The SHAP summary plot is a visualisation used to show the contribution of each feature to the model’s prediction results. In the SHAP summary plot, the SHAP value of each feature is typically displayed, along with the direction of the feature’s impact on the prediction results (positive or negative). By observing the SHAP summary plot, one can intuitively understand the importance of each feature to the final prediction results, as well as how changes in feature values affect the prediction results. This study revealed several key variables related to the risk of AKI occurrence in patients through the SHAP summary plot. Among these, an increase in urine output is associated with a decreased risk of AKI, while an increase in body weight is associated with an increased risk of AKI. Increases in white blood cells, neutrophils, serum creatinine, blood urea nitrogen, PT and PTT values are associated with an increased risk of AKI. Additionally, a decrease in minimum systolic blood pressure is associated with an increased risk of AKI occurrence in patients. The use of vasopressors and mechanical ventilation is associated with an increased risk of AKI. Avoiding the use of antibiotics is associated with a reduced risk of AKI in patients. Higher GCS scores are associated with a decreased risk of AKI occurrence in patients.
Low blood pressure is considered essential for organ perfusion; therefore, hypotension is associated with poor prognosis. Currently, most studies focus on the relationship between mean arterial pressure (MAP) and AKI. A prospective observational study29 demonstrated that patients who developed AKI had significantly lower time-adjusted MAP compared with those who did not progress to AKI (74.4 mm Hg vs 78.6 mm Hg, p<0.001). An MAP below 73 mm Hg was identified as an independent predictor of AKI progression. Low MAP may be insufficient to protect renal function, while elevated MAP is associated with improved tubular function and lower serum creatinine levels.30 An experimental study31 revealed that animals in the low MAP group had higher median plasma creatinine levels than those in the high MAP group, with AKI incidence rates of 50% and 38% respectively at 12 hours post-untreated sepsis. Almost all studies suggest that the kidneys’ autoregulatory capability is impaired under low MAP conditions, leading to inadequate renal perfusion. Furthermore, in cases of severe infection or shock, changes in microcirculation and reduced vascular reactivity may raise the autoregulatory threshold of the kidneys in response to MAP. Baek et al 32 explored the optimal SBP range for patients with acute kidney injury in a retrospective study, finding a U-shaped relationship between SBP and the severity of AKI or 90-day mortality within 48 hours after AKI onset, indicating that both low and high blood pressures may have adverse effects. We propose that the kidneys are highly sensitive organs regarding blood flow perfusion, and early sepsis patients often exhibit hypotension, affecting the glomerular filtration rate. Systemic vasodilation and capillary leakage can reduce effective circulating blood volume, further exacerbating hypotension and organ perfusion deficits, thereby triggering or worsening AKI. Although these haemodynamic changes activate the renin-angiotensin system, leading to renal vasoconstriction, this constriction may be insufficient to entirely compensate for reduced blood flow in the early stages. Moreover, early decreases in systolic blood pressure may worsen the microcirculatory dysfunction caused by the systemic inflammatory response in sepsis, adversely affecting the supply of oxygen and nutrients to the kidneys, further aggravating renal injury.
Patients with pancreatitis complicated by sepsis often require vasopressor support, as fluid therapy alone is insufficient to correct the systemic vasodilation and endothelial dysfunction induced by sepsis.33 Additionally, septic patients typically have low urine output, and aggressive fluid resuscitation or diuretic therapy can lead to an increased risk of fluid retention. Fluid overload is associated with poorer patient outcomes; in a retrospective study, Legrand et al 34 identified a correlation between new-onset or persistent AKI and elevated central venous pressure (CVP), with venous congestion induced by fluid resuscitation increasingly recognised as a contributing factor to renal injury. Norepinephrine is the first-line agent for patients with sepsis, and its increasing dosage has been linked to a higher incidence of AKI progression, potentially due to excessive vasoconstriction in regional vascular beds. Furthermore, some studies indicate that the use of diuretics may be associated with an increased risk of AKI. Loop diuretics can inhibit sodium reabsorption in the macula densa, thereby stimulating the renin-angiotensin-aldosterone system (RAAS) and leading to AKI, while some cases of AKI may stem from the combined effects of diuretics and other medications, including antibiotics, contrast agents and ACE inhibitors/ARBs.35 Thus, appropriate fluid resuscitation can correct fluid losses and improve microcirculation and tissue oxygenation.
Previous studies have reported that the incidence of AKI within the first 48 hours of mechanical ventilation ranges from 15.5% to 17.1%.36 In the context of the pathophysiology of sepsis, factors associated with AKI include impaired gas exchange and severe hypoxemia. Mechanical ventilation may lead to haemodynamic changes,37 such as hypotension and fluid-responsive shock, which affect tubular perfusion and decrease glomerular filtration rate (GFR) by reducing cardiac output and stimulating hormonal and sympathetic nervous responses, ultimately resulting in AKI. Increasing evidence suggests that the pro-inflammatory effects of positive pressure ventilation (PPV) may be a contributing factor to AKI. Douillet et al 38 demonstrated that mechanical ventilation can alter the expression of nucleotide and purinergic receptors in the kidneys, and inappropriate mechanical ventilation strategies can induce the production of various inflammatory cytokines (such as IL-8 and monocyte chemotactic protein), leading to apoptosis of renal epithelial cells.
Dysregulation of the immune system and the release of inflammatory factors are direct pathophysiological mechanisms underlying sepsis-related kidney injury.39 Abnormal white blood cell counts, reflecting cellular immune dysregulation, may exacerbate the risk of AKI. Our study indicated that white blood cell count is a risk factor for the occurrence of AKI in sepsis patients, consistent with previous research. Elevated white blood cell counts indicate the body’s response to infection but also signify the persistence of the inflammatory process.40 The release of inflammatory mediators may have direct toxic effects on the kidneys. When white blood cells become activated or accumulate in the microvasculature, this can further obstruct microcirculation, exacerbating renal hypoxia and damage, thereby promoting the onset of AKI.
The use of antibiotics in sepsis patients may lead to renal toxicity, particularly with certain classes such as aminoglycosides and β-lactams. Therefore, avoiding their use can alleviate renal burden, especially in cases where renal function is already compromised due to infection. Patients who do not receive antibiotics may, to some extent, maintain their immune balance, relying on their immune response to address mild or early infections, which can help reduce systemic inflammation and decrease the risk of AKI. Furthermore, antibiotics may disrupt the gut microbiome balance, and a healthy microbiome is crucial for regulating immune responses and combating infections. Patients not receiving antibiotics typically exhibit milder infection symptoms, which may correlate with other favourable baseline characteristics and a lower risk of AKI. It is important to note that the observed association based on SHAP values from model visualisation does not necessarily imply that antibiotic use directly leads to an increase in AKI. Our conclusions primarily suggest a potential relationship between the two, rather than a direct causal link. Future research should focus on the use of specific antibiotics and their specific impact on the risk of AKI, as well as a deeper exploration of the underlying mechanisms.
In summary, weight gain is associated with an increased risk of AKI, therefore, clinicians should develop personalised fluid management plans based on individual patient characteristics. In particular, for obese patients, careful assessment of fluid intake and medication dosages is necessary to avoid excessive fluid overload and the subsequent risk of AKI. Physicians should regularly monitor blood pressure in septic patients and, if necessary, take timely measures for fluid resuscitation and the use of vasoactive medications to ensure adequate renal perfusion. When using diuretics, it is essential to carefully evaluate their necessity, especially in patients with insufficient fluid load, and prioritise alternative management strategies to avoid impacting renal function. Given that fluid overload is associated with poor outcomes in AKI, fluid therapy should involve individualised management, with regular assessments of the patient’s fluid status. In particular, after large-volume fluid resuscitation, ongoing monitoring of central venous pressure (CVP) and urine output is crucial for the timely detection and correction of fluid overload. If mechanical ventilation is required, attention should be paid to the choice of ventilation strategy to optimise ventilation modes and minimise potential adverse effects on the kidneys while enhancing haemodynamic management. Additionally, monitoring of the inflammatory status in septic patients should be strengthened to promptly identify bacterial infections and inflammatory responses and to develop effective anti-inflammatory strategies. Given the association between AKI and coagulopathy, enhanced monitoring of coagulation function is needed, along with heightened vigilance regarding the risk of bleeding.
Our model successfully predicted whether patients would experience AKI during their ICU stay. This study provides strong support for identifying the likelihood of AKI occurrence in AP patients with concomitant Sepsis, aiding in better prevention and management of such complications in clinical practice.
In this study, there are several noteworthy limitations. (1) Given the use of a large public database with many missing values in patient medical records, the performance of our model largely depends on the accuracy of data recording, which may introduce some degree of bias. (2) While feature selection was performed during model construction to reduce complexity and improve generalisation, and L1 and L2 regularisation methods were introduced to prevent overfitting, in the external validation cohort, although the AUC value of the ensemble model was relatively high, it did not reach the optimal level. This could be attributed to the smaller sample size and poorer data quality during both model construction and external validation. (3) Theoretically, all sepsis patients should receive antibiotic treatment, however, in the MIMIC-IV database, not all patients received antibiotics, leading to potential discrepancies from theoretical expectations, while the lower antibiotic usage rate in the MIMIC-III database can be partially attributed to its smaller sample size. (4) The model still has certain shortcomings in explaining the pathophysiology of disease occurrence. (5) Due to the inability to obtain patients’ BMI data, there may be some differences in the study conclusions compared with existing research.
Conclusion
The stacked ensemble model, named ‘Multimodel,’ developed in this study, achieved AUC values of 0.853 and 0.802 in internal and external validation cohorts, respectively. It performed excellently on other metrics as well, making it a reliable tool for predicting AKI in patients with acute pancreatitis complicated by sepsis. Additionally, the SHAP model explanation method aids physicians in understanding and evaluating prediction results, thus, facilitating the development of personalised treatment plans.
Data availability statement
Data are available upon reasonable request. Datasets generated and analysed during the current study may be obtained upon reasonable request.
Ethics statements
Patient consent for publication
Ethics approval
The establishment of the MIMIC database was approved by the Massachusetts Institute of Technology (Cambridge, MA) and Beth Israel Deaconess Medical Center (Boston, MA), and consent for original data collection was obtained. Therefore, in the research in this database, we waived the need for ethical approval statements and informed consent.
References
Footnotes
Contributors FL analysed the data and wrote the paper, as the first author. ZW extracted data from the database and assisted in constructing models. RB checked the integrity of the data and the accuracy of the data analysis. ZX and JC participated in the writing and revision of the article. FL, ZW and YZ jointly designed and revised this article. All authors read and approved the final manuscript. ZW is responsible for the overall content as guarantor. The authors would like to thank the Massachusetts Institute of Technology and the Beth Israel Deaconess Medical Center for their support of the MIMIC project.
Funding This work was supported by the (National Natural Science Foundation of China) Grant number (82160131) and (The Department of Science and Technology in Qinghai Province, China) Grant number (2021-ZJ-963Q).
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.