Article Text
Abstract
Introduction The TransEAsome project, funded by the Agence Nationale de la Recherche, aims to evaluate the long-term outcomes of patients with oesophageal atresia (OA) between 13 and 14 years old and establish multiomics profiles using data from the world’s biggest OA registry.
Methods and analysis TransEAsome is a national multicentre population-based cohort study recruiting participants from all qualified French centres for OA surgery at birth. The primary objective is to assess the prevalence of gastro-oesophageal reflux disease in adolescence among patients with OA, with several secondary objectives including the identification of risk factors and multiomic profiles from oesophageal biopsies and blood samples collected between 13 and 14 years old, compared with a control group. This comprehensive characterisation of phenotype and omic profiles aims to enhance the understanding of disease evolution in patients with OA and inform tailored care management strategies.
Ethics and dissemination The study, coconstructed with input from patients, parents and research-expert adolescents, has obtained approval from the ethics research committee: Comité de protection des personnes Est II. Findings will be disseminated to various target audiences, including the scientific community, research participants, the patient community, the general public, regulatory authorities and policymakers. Data will be made available in a Findable, Accessible, Interoperable, Reusable format on the France Cohortes platform on study completion.
Trial registration number NCT05995171:Clinical trial
- Adolescents
- Oesophageal disease
- Patient Reported Outcome Measures
- Quality of Life
- Paediatric gastroenterology
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
- Adolescents
- Oesophageal disease
- Patient Reported Outcome Measures
- Quality of Life
- Paediatric gastroenterology
STRENGTHS AND LIMITATIONS OF THIS STUDY
Multicentre, national, population-based cohort study conducted in all French centres qualified to perform oesophageal atresia surgery at birth.
Longitudinal analysis facilitated by a nested cohort design. The birth and 1-year-old visits are in the national register of oesophageal atresia (Registre National de l’ATresie de l’Oesophage (ReNATO)), the 6-year-old visits into the nested cohort COMAD6 (Cohorte nationale sur les Maladies de l’Appareil Digestif), while TransEAsome focuses on the 13 and 14-year-old visits,
Active involvement of patients and parents thanks to the users’ representative committee, ensuring the study reflects patient perspectives.
Comprehensive multiomic analysis encompassing transcriptomic, epigenetic, proteomic and metabolomic data, providing a holistic understanding of the disease.
Omic studies rely on oesophageal tissue samples collected for clinical surveillance purposes, which may not be systematic, and long-term storage could lead to RNA degradation over time, potentially impacting data quality.
Introduction
Oesophageal atresia (OA) is a rare congenital anomaly affecting approximately 160 newborns annually in France, characterised by a discontinuity in the oesophagus. It is often associated with other malformations in over 50% of cases.1 Since the early 1950s, significant advancements in surgical techniques have vastly improved the management of this condition, resulting in a favourable prognosis. While the mortality rate has declined to less than 7% in developed nations like France,2 morbidity remains noteworthy.3 4
Gastro-oesophageal reflux disease (GORD) is more prevalent among individuals with OA compared with the general population.5 Studies indicate that approximately 30% of patients with OA experience GORD at least once in their lifetime.6 Complications of GORD, such as peptic esophagitis and Barrett’s oesophagus, present heightened risks for oesophageal cancer,7 with documented cases of adenocarcinoma or squamous cell carcinoma in young adults with OA.8
Dysphagia is a common concern among patients with OA, with around 45% of 5-year olds experiencing swallowing difficulties attributed to anastomotic strictures or oesophageal dysmotility.9 Eosinophilic oesophagitis (EoE), a rising concern in oesophageal health, has been noted to occur more frequently in patients with OA, contributing to dysphagia.10
Challenges in eating often lead to undernutrition, affecting up to one-third of children with EA by the age of five.11 Respiratory complications are also prevalent,12 particularly in the early years, significantly impacting long-term quality of life (QoL).13
To better understand the long-term outcomes and predict complications, our objective is to investigate a nested population cohort of adolescents born with OA. Leveraging the existing national population-based registry (ReNaTo), and the nested clinical cohort, COMAD6, consisting of 382 children born between 2010 and 2012, we aim to compile a new database comprising clinical data, blood samples and oesophageal biopsies obtained during routine follow-up between 13 and 14 years old (figure 1).
Follow-up of a nested cohort of patients with OA born between 2010 and 2012. OA, oesophageal atresia.
This nested cohort, within the world’s largest registry of more than 2300 patients with OA, offers a unique opportunity to answer questions about outcomes of OA at adolescence in terms of morbidity and health status, to improve care and follow-up and prevent long-term complications.
In addition to the phenotypic assessment, our investigations extend to exploring the biological mechanisms underpinning complication development through genomic, transcriptomic, proteomic, epigenetic and metabolomic analyses of the oesophagus.
While upper gastrointestinal endoscopy (EGD) is standard in the care of symptomatic or not patients with OA,5 its invasive nature necessitates hospitalisation and general anaesthesia in children.
To date, there is little data on the epigenetic, genomic14–18 or transcriptomic19 profile of patients with OA. In a small study of 10 children with Barrett’s oesophagus (some of them operated at birth for OA), the authors performed fluorescence in situ hybridisation with probes on 4 micron sections taken from sequential paraffin-embedded biopsies and identified four probe sets reported to be associated to adult Barret adenocarcinoma.19 Genetic markers were also identified in adult Barrett’s adenocarcinoma patients. This preliminary study shows that, even at an early age, Barrett’s may show genetic changes associated with neoplastic progression.20 Another recent study examined the relationship between eosinophilic esophagitis (EoE) and OA by profiling the transcriptional signature of EoE.21 Using an in silico approach, they found six genes differentially expressed between the two entities (OA+ EoE+ and OA− EOE+). Two of them were associated with major dysphagia, the development of strictures, and the need for dilatations in patients with OA+EoE+.
As EGD and oesophageal biopsies are invasive procedures that usually require hospitalisation and general anaesthesia in children, using a more accessible biological sample as blood plasma will also be investigated to see if the markers of the oesophagus translate into blood.
The overarching goal of TransEAsome is to construct a vast longitudinal database including phenomenal and exposomal patient data amalgamating clinical, biological, environmental and lifestyle data alongside multimodal omics analyses: RNA abundance, protein abundance, proteogenomic profiling, protein modification, metabolite abundance, methylated sites. This innovative approach holds promise for enhancing the prediction of adult OA outcomes, including the identification of risk factors for future health complications such as oesophageal cancer and EoE.
Objectives
TransEAsome will address the evolution of patients with OA at the time of adolescence by establishing a unique and comprehensive database that integrates clinical and omics data in a structured and interoperable format to evaluate their health status and QoL. This project is designed to achieve the following objectives:
Evaluate the prevalence of GORD during adolescence in the OA population.
Identify factors associated with GORD during adolescence in the OA population.
Assess the QoL, nutritional status and frequency of respiratory complications in the OA population.
Compare the single and multiomic profiles derived from oesophageal biopsies in adolescents operated at birth for OA with those who did not, aiming to delineate alterations in biological pathways specific to OA.
Investigate changes in the expression of omics variables within the identified biological pathways over time in the OA population, utilising data obtained from EGD and biopsies.
Evaluate the correlation between the expression levels of microRNAs and proteins in plasma compared with oesophageal biopsies in the OA population, utilising data from individuals who underwent EGD, biopsies and blood sampling at ages 13 and 14.
Methods and analysis
Recruitment sources
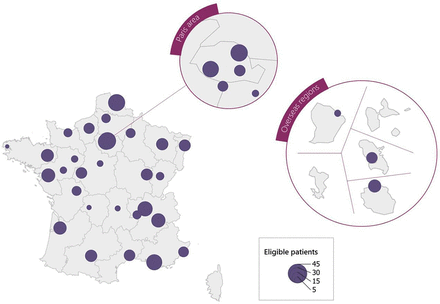
Patients for the TransEAsome study will be recruited from the French national registry for OA known as ‘ReNaTo’, which involves all 35 OA competence centres across France. As of the conclusion of 2022, more than 2300 patients were included in ReNaTo. Among them, 492 are projected to reach 13–14 years of age during the recruitment period of the TransEAsome project. However, an anticipated loss to follow-up necessitates accounting for a pool of 300 patients. Leveraging the national register ReNaTo, the project management team will furnish each centre with a pseudo-anonymised patient roster, facilitating the identification of eligible candidates from their patient cohorts. Given the unique demographic characteristics of each participating centre, recruitment potentials vary, spanning from 1 to 34 patients per centre during the study period (figure 2).
Localisation of recruitment in France.
For the objectives involving multiomic profiling, the control group will be drawn from six selected centres. Eligible patients will be identified through the extraction of cases meeting the study’s inclusion criteria from the pathology department.
Study population
TransEAsome eligibility and exclusion criteria are listed in table 1. The initial recruitment period will start in September 2023 and end in May 2026. Our goal will be to enrol 300 patients and 150 controls.
Eligibility and exclusion criteria
The blood substudy as well as the inclusion of controls will only be performed in Lille, Paris Necker, Paris Robert Debré, Lyon, Grenoble and Marseille for logistic, feasibility and cost-saving reasons.
Patient and public involvement
A young patient advisory committee and a parents/patient representativeness committee have been established to ensure the active involvement of young patients and their parents throughout all phases of the TransEAsome project, from inception to dissemination of results. These committees have played a pivotal role in shaping the study protocol, including determining the data to be collected and reviewing informed assents and consents before regulatory submissions. Throughout the trial, these committees will contribute to the decision-making process of the steering committee and provide insights from a patient perspective on the conduct of the study.
The committees will remain actively engaged throughout the project, facilitating communication and dissemination efforts to the scientific community, patients and the public regarding key milestones, actions and emerging knowledge. On project completion, the committees will review the analysis, discuss the findings and aid in disseminating results.
Kids France (Hospices Civils de Lyon, Pedstart) will oversee the patient and public involvement (PPI) activities in collaboration with the French patient with OA support group. Kids France will ensure that PPI activities are not only meaningful for the research but also beneficial for the patients themselves. Training related to PPI activities will be provided to the committees by Kids France. Reporting activities to the steering committee, feedback on suggestions from patients and parents and assessment of impact are integral components of the TransEAsome PPI process.22 23
Outcomes and assessments
Outcomes and assessments are presented in with separate forms for parents and adolescent patients (table 2).
Outcomes and assessments between 13 and 14-year olds
QoL questionnaires and blood sampling are the only assessments that are not part of routine practice. Both the generic paediatric QoL assessment (Pediatric Quality of Life Inventory (PedsQL) and the disease-specific QoL questionnaire (EA QoL)13 will be administered in this cohort, with separate forms for parents and adolescent patients.
Biological samples
Oesophageal tissue
EGD and biopsy sampling can be conducted at any point during the follow-up of patients with OA, based on symptoms and routine evaluation.24 Patients who undergo these procedures will be requested to authorise the collection of four sections of formalin-fixed paraffin-embedded (FFPE) oesophageal tissue. These samples are stored in the pathology departments for a minimum of 10 years, per French regulations (décret N°88–280 du 24 mars 1988 de l'article L.761–11 du code de la santé publique) and are commonly stored for approximately 25 years.
FFPE storage is the established standard method for the long-term preservation of biological tissue in routine care, universally practised across all participating centres. Whenever feasible (subject to patient authorisation and technical feasibility), previously collected oesophageal biopsies from included patients in routine care will also be analysed or pooled for analysis. FFPE sections sourced from all participating centres will be centralised at the Biological Resource Centre of Lille University Hospital (CRB du CIC 1403 du CHU de Lille, BRIF BB0033-00030) until procurement for analysis.
Comparative analysis of omic profiles measured at different time points for the same patient will facilitate longitudinal comparisons. The objective is to include biopsies from 150 patients in the analysis.
Plasma
Plasma collection will be conducted at selected centres following consultations for adolescents who have available oesophageal biopsies. Whole blood will be drawn into a 5 mL EDTA tube between the ages of 13 and 14 years and subsequently centrifuged. Plasma aliquots will then be stored at −80°C and centralised at the Biological Resource Centre of Lille University Hospital (CRB du CIC 1403 du CHU de Lille, BRIF BB0033-00030) until procurement for analysis.
Standard Operating Procedures for blood and biopsy collection, storage and shipping will be uniform across all participating centres to ensure consistency and quality control.
Data sources, collection and monitoring
Clinical data collection
Clinical data will be recorded in both a traditional paper-based case report form (CRF) and an electronic CRF using the Ennov Clinical platform. Monitoring of completion will be conducted, and participants will receive reminders to complete QoL questionnaires if necessary.
The study will adhere to the guidelines set forth by the International Conference on Harmonization (ICH) Good Clinical Practice (GCP) outlined in ICH GCP Topic E6 (1996), ensuring compliance with ethical and quality standards in clinical research.
Omics data generation
Omics data will be generated from both oesophageal mucosa and plasma samples.
For proteomics analysis, FFPE tissues and plasma extracellular vesicles (EVs) will undergo antigen retrieval before localised trypsin digestion of the regions of interest. The resulting peptides will be collected using a liquid micro junction before shotgun analyses. In parallel, total RNA extraction will be performed to prepare next-generation sequencing 3'RNA-seq and small RNA-seq (sRNA-seq) libraries with molecular barcodes (unique molecular identifiers—UMI). The incorporation of UMIs in the sequencing libraries will enable accurate quantification even in cases of low RNA yield and quality. The sRNA-seq libraries will facilitate the detection and quantification of non-coding RNA such as microRNA (miRNA) and small nucleolar RNA (snoRNA). All libraries will be sequenced using a NovaSeq 6000 instrument.
For plasma proteomics analysis, EVs will initially be isolated using size exclusion chromatography. Fractions will be collected and assessed for size and concentration using Nanosight analysis. EV-positive fractions will then be pooled before enzymatic digestion of proteins and subsequent identification by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS). RNA extraction from EVs in the pooled fractions will be performed using the Direct-zol RNA extraction kit, followed by analysis as described above.
Omics data preprocessing
The protein identification process will use MaxQuant software, comparing all MS/MS data with the human protein database from the Uniprot bank. Key parameters in MaxQuant will be defined, including trypsin as the digestion enzyme with a maximum of two missed cleavages, methionine oxidation and N-terminal protein acetylation as variable modifications, and carbamidomethylation of cysteines as a fixed modification. Label-free quantification (LFQ) will be performed using default settings. Initial mass tolerance will be set to 6 ppm for MS mode and 20 ppm for fragmentation data in MS/MS mode. Protein and peptide identification parameters will be configured with an FDR (false discovery rate) of less than 1%, requiring a minimum of two peptides per protein, including at least one unique peptide.
Preprocessing analysis will commence using Perseus software. LFQ intensity values of each sample will be imported from MaxQuant into Perseus, and the data matrix will undergo contaminant removal. Subsequently, the data will be log2 transformed, and a filter will be applied to retain proteins present in at least 70% of samples within at least one group.
The RNA-seq analysis pipeline will involve bclconverter for fastQ generation, fastp for trimming and UMI first-step processing, fastQC for quality control, STAR for alignment, umi-tools for deduplication, qualimap for alignment quality control and featureCounts for quantitative assessment of genes. The sRNA-seq analysis pipeline will follow a similar workflow to RNA-seq, utilising bowtie for alignment with necessary adjustments.
For methylation analysis, the pipeline will include bclConverter, fastQC, trim_galore, additional python scripts, bismark25 and bowtie2 for alignment and deduplication, with qualimap used for alignment QC.
Statistical analysis
Sample size
The rationale for the number of patients was determined based on the cohort of patients included in the OA registry who will be between 13 and 14 years old during the recruitment period (n=492). Accounting for potential dropouts, refusals to participate and deaths, it is estimated that approximately 300 patients can be included, representing around 30% of those eligible. This estimation aligns with the observed rate of non-inclusion of 22% observed in the COMAD6 study.
With a cohort of 300 patients, the study will have the capacity to estimate the frequency of GORD in adolescents (primary objective) with a maximum absolute precision of 5.7%. This precision is represented by the half-width of the 95% CI, particularly in scenarios where the observed frequency of GORD is 50%, which presents the greatest width of the CI for an equivalent population size.
Regarding the control group without OA, the plan is to include 150 patients, taking into consideration logistical and financial constraints associated with various omics analyses.
For the blood substudy, participating centres have a potential recruitment capacity of 176 patients with OA (ranging from 20 to 30 per centre). Anticipating losses to follow-up and refusals, it is estimated that approximately 150 patients can be included in this sub-study.
Phenotyping
Frequency of GORD
The frequency of patients born with a diagnosis of OA and GORD in adolescence will be calculated along with its two-sided 95% CI. The CI will be estimated using the normal approximation method.
Here is the general formula to calculate the CI for a proportion (in this case, the frequency of patients with OA and GORD):

where:
Frequency: proportion of patients with OA and GORD.
Z: Z-score corresponding to the desired confidence level (for a 95% CI, Z ≈ 1.96).
Sample size: total number of patients in the study.
Given the calculated proportion of patients with OA and GORD and the total sample size, we can plug these values into the formula to obtain the CI.
Associated factors with GORD
To assess the independent factors associated with the presence of GORD in adolescence, the following methodology will be employed:
Bivariate logistic regression models:
Bivariate logistic regression models will be conducted to evaluate the ORs of GORD and their corresponding 95% CIs. These models will assess the relationship between each individual factor and the presence of GORD.
For quantitative factors, the log-linearity assumption will be assessed using restricted cubic spline functions26
Selection of factors for multivariable analysis:
Factors associated with the presence of GORD with a significance level of less than 0.20 in bivariate analyses will be included in a multivariable backward-stepwise logistic regression model.
If the number of events per candidate variable is insufficient (<10), a penalised method will be used.
Collinearity assessment:
Collinearity between the candidate factors for multivariate analysis will be examined by calculating the variance inflation factor. An alert threshold will be set at 2.5.
Development and evaluation of multivariable model:
Discrimination of the selected multivariable model, indicating its ability to differentiate between patients with and without GORD, will be assessed using C-statistics corrected for over-optimism via bootstrap resampling.26
Calibration, which measures the agreement between predicted and observed probabilities of GORD, will be evaluated using the Hosmer-Lemeshow goodness of fit test.
Handling data:
To prevent case deletion due to missing data in multivariable analysis, missing data will be imputed.
Simple imputation will be used if the missing data rate is below 10%, while multiple imputations27 will be employed otherwise.
The number of imputations (m) will be determined based on the fraction of missing information.
Imputations will be performed using the Multiple Imputation by Chained Equations procedure, incorporating all variables included in the analyses.28 Missing data on quantitative variables will be imputed using the predictive mean matching method, while those on qualitative variables will be imputed using logistic regression models (binomial, ordinal or multinomial, depending on the number and order of modalities). In the case of multiple imputations, Rubin’s rules will be applied to combine estimates obtained from each imputed dataset29 in the case of multiple imputations.
Health status
The health status and QoL assessment criteria in adolescence will be described using the following parameters:
QoL scores:
QoL scores obtained from the PedsQL and the EA-QoL questionnaires will be reported.
Anthropometric measures:
Weight
Height
Weight/height Z-score
BMI Z-score.
Frequency of respiratory complications
For each of these criteria, positional parameters, such as means or medians, will be calculated, and their two-sided 95% CIs will be reported.
The CIs will provide a range of plausible values for these parameters, allowing for an understanding of the precision of the estimates. These intervals will be calculated using appropriate statistical methods, such as the t-distribution for means or the bootstrap method for medians, depending on the distributional characteristics of the data.
Omic analysis
Omic profile between 13 and 14 years
For each type of omics data, differential analyses between the group with OA and the group without OA will be conducted using appropriate R packages. For example:
Proteomic data will be analysed using the limma package.30
Sequencing data will be analysed using DESeq2.31
Methylation levels extracted from bismark will be analysed using packages such as methylKit32 and RnBeads.33
Once differential analyses are performed, the impacted metabolic pathways will be identified through enrichment analyses. This can be accomplished using R packages like ClusterProfileR.34
Further classification analysis will be conducted using R packages such as methylClass35 to study the evolution of omic profiles over time.
For each identified metabolic pathway, differences in means of corresponding gene expression, methylation or protein abundance between two time points will be tested using parametric or non-parametric tests, depending on the data distribution. There will be a particular focus on the biological pathways of EoE and Barrett’s oesophagus. Parametric tests will use similar approaches as for the initial differential analyses, while non-parametric tests such as Wilcoxon signed-rank tests may be employed.
To account for multiple testing, the p values of these tests will be corrected. The Bonferroni procedure may be used initially to control the family-wise error rate, followed by the Benjamini-Hochberg procedure to control the false discovery rate if necessary. These corrections will help ensure the reliability of the identified associations and reduce the likelihood of false positives.
Transposition from the biopsies to the blood
Scatterplots depicting microRNA expression and protein abundance in plasma will be generated, with levels in biopsies serving as the independent variable. These scatterplots will visually illustrate the relationships between the variables. Additionally, correlation coefficients will be calculated to quantify the strength and direction of these relationships, providing numerical measures of association.
Linear regression analysis will be conducted to further explore the relationships between microRNA expression, protein abundance in plasma and levels in biopsies. This analysis will involve fitting a linear model to the data to determine the extent to which changes in one variable predict changes in another. The residuals from the regression model will be examined to assess the adequacy of the model fit and identify any potential outliers or patterns in the data that may require further investigation.
Overall, these analyses will provide insights into the relationships between microRNA expression, protein abundance in plasma and levels in biopsies, helping to uncover potential biomarkers or indicators of interest in the context of OA.
Ethics and dissemination
The study (protocol V.1.1, 2.0, 3.0 and 4.0) has obtained approval from the French ethics research committee (Comité de protection des personnes Est II).
Informed consent
Patients and their parents will receive prior communication before the scheduled follow-up visit, providing them with information about the opportunity to participate in the study. The study protocol will be thoroughly explained, and an information note will be sent to them to allow ample time for consideration, with a minimum duration of 1 week.
On the day of the visit, any queries or concerns they may have will be addressed comprehensively. Subsequently, they will have the option to confirm or decline their participation in the study, based on their informed decision.
For the control group, eligible patients will be contacted to invite their participation in the study. A letter of non-opposition will be sent to obtain their agreement for the utilisation of samples collected during routine care, which will be repurposed for research purposes.
Privacy
The TransEAsome team will prioritise the pseudo-anonymisation of all clinical and omics data, ensuring that individual identities are solely identifiable by the patient inclusion number. Access to the data will be restricted, with each partner granted limited access based on their specific requirements for task performance.
The project will adhere to the guidelines established by the Commission Nationale de l’Informatique et des Libertés, the French data protection authority, particularly following standard methodology 001 for clinical research involving the collection of patient consent. These measures aim to uphold patient confidentiality and privacy while facilitating valuable research endeavours.
Dissemination plan
Data
All data collected will undergo harmonisation with standard terminologies encompassing pathologies, localisation, treatments, units of measure, phenotypes and health interventions. This harmonisation process aims to enhance interoperability with other databases and enable seamless reuse by diverse research teams.
The data will be centralised within the France Cohortes platform, where each entry will be assigned a unique identifier. Additionally, metadata files will be generated per Findable, Accessible, Interoperable, and Reusable (FAIR) principles, ensuring that the data remain FAIR.
Results
The dissemination of findings from the TransEAsome project will target various audiences to ensure broad impact and relevance. These audiences include:
Scientific community: findings will be disseminated through peer-reviewed scientific publications and conference presentations, allowing researchers and clinicians to access and use the latest advancements in the field.
Research participants: participants in the study will be informed of the results through participant newsletters, ensuring that they remain informed about the outcomes of the research to which they contributed.
Patient associations: results will be shared with patient associations to empower patients and their families with knowledge about advancements in the understanding and management of OA.
General public: information about the project’s findings will be communicated to the general public through social media platforms such as Twitter and LinkedIn, raising awareness and understanding of OA among the wider community.
Regulatory authorities and policymakers: relevant findings will be shared with regulatory authorities and policymakers to inform decision-making processes and contribute to the development of policies related to rare diseases and paediatric healthcare.
Communication efforts will adhere to the graphic charter of the project, ensuring consistency and professionalism in visual presentation. Additionally, all communications will acknowledge the funding provided by the Agence Nationale de la Recherche, as required. This comprehensive dissemination strategy aims to maximise the impact of the TransEAsome project across various stakeholders and promote the translation of research findings into tangible benefits for patients and society.
Ethics statements
Patient consent for publication
Acknowledgments
The authors express their gratitude to the scientific board of the TransEAsome project for their invaluable input in shaping its design. They also extend their appreciation to all investigation teams for their significant contributions to the establishment and execution of this study. Special thanks are extended to Katialine Groff and Dr. Rony Sfeir for their dedicated work on the oesophageal atresia national registry ReNaTo, which has been instrumental in the project's progress. The authors also acknowledge the members of the user’s committees for their insightful feedback, which has consistently enriched the project. Additionally, they express their gratitude to the paediatric clinical investigation centre at Lille Hospital for their technical support in clinical research, which has been indispensable to the authors. This collaborative effort and support from various individuals and institutions have been essential in advancing the TransEAsome project and its objectives.
References
Footnotes
Collaborators University Hospital of Amiens, University Hospital of Angers, University Hospital of Besançon, University Hospital of Bordeaux, University Hospital of Brest, University Hospital of Caen, Public Hospital of Cayenne, University Hospital of Clermont Ferrand, University Hospital of Dijon, University Hospital of Fort de France, University Hospital of La Tronche, Public Hospital of Le Mans, University Hospital of Limoges, University Hospital of Lyon, University Hospital of Marseille, University Hospital of Montpellier, University Hospital of Nancy, University Hospital of Nantes, University Hospital of Nice, Public Hospital of Orléans, University Hospital of Paris Armand Trousseau, University Hospital of Kremelin Bicêtre, University Hospital of Créteil, University Hospital of Paris Necker, University Hospital of Paris Robert Debré, University Hospital of Poitiers, University Hospital of Pointe à Pitre, University Hospital of Reims, University Hospital of Rennes, University Hospital of Rouen, University Hospital of Saint Denis de la Réunion, University Hospital of Saint Etienne, University Hospital of Strasbourg, University Hospital of Toulouse and University Hospital of Tours.
Contributors FG is the scientific coordinator and guarantor. MF, RH, GM, MS are partner leaders in the project. MA is the Lille University Hospital’s principal investigator, JV is the co-technical-head of the Billile platform, SG is head of the patient and public involvement task, JL is the head of the Lille University Hospital’s biostatistics department, LD is a methodologist who revised the protocol submitted to ethics committee, MD is in charge of the omics analysis within the PRISM team, SF is a head of the ReNaTo registry, JR and VA are a members of the French national patient with OA association and ML is the project manager. All authors have contributed to the writing of the study protocol and have approved the final version of this manuscript.
Funding This project has received funding from the ANR program France 2030 under the specific grant agreement ANR-21-PMRB-0011. As well as a funding from the Institution de prévoyance et Retraite Collective des Employés de Maison (IRCEM) fundation.
Map disclaimer The inclusion of any map (including the depiction of any boundaries therein), or of any geographic or locational reference, does not imply the expression of any opinion whatsoever on the part of BMJ concerning the legal status of any country, territory, jurisdiction or area or of its authorities. Any such expression remains solely that of the relevant source and is not endorsed by BMJ. Maps are provided without any warranty of any kind, either express or implied.
Competing interests None declared.
Patient and public involvement Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review Not commissioned; externally peer-reviewed.