Article Text
Abstract
Introduction Children represent a large and vulnerable patient group. However, the evidence base for most paediatric diagnostic and therapeutic procedures remains limited or is often inferred from adults. There is an urgency to improve paediatric healthcare provision based on real-world evidence generation. Digital transformation is a unique opportunity to shape a data-driven, agile, learning healthcare system and deliver more efficient and personalised care to children and their families. The goal of Paediatric Personalized Research Network Switzerland (SwissPedHealth) is to build a sustainable and scalable infrastructure to make routine clinical data from paediatric hospitals in Switzerland interoperable, standardised, quality-controlled, and ready for observational research, quality assurance, trials and health-policy creation. This study describes the design, aims and current achievements of SwissPedHealth.
Methods and analysis SwissPedHealth was started in September 2022 as one of four national data streams co-funded by the Swiss Personalized Health Network (SPHN) and the Personalized Health and Related Technologies (PHRT). SwissPedHealth develops modular governance and regulatory strategies and harnesses SPHN automatisation procedures in collaboration with clinical data warehouses, the Data Coordination Center, Biomedical Information Technology Network, and other SPHN institutions and funded projects. The SwissPedHealth consortium is led by a multisite, multidisciplinary Steering Committee, incorporating patient and family representatives. The data stream contains work packages focusing on (1) governance and implementation of standardised data collection, (2) nested projects to test the feasibility of the data stream, (3) a lighthouse project that enriches the data stream by integrating multi-omics data, aiming to improve diagnoses of rare diseases and 4) engagement with families through patient and public involvement activities and bioethics interviews.
Ethics and dissemination The health database regulation of SwissPedHealth was approved by the ethics committee (AO_2022-00018). Research findings will be disseminated through national and international conferences and publications in peer-reviewed journals, and in lay language via online media and podcasts.
- Child
- Electronic Health Records
- EPIDEMIOLOGY
- PAEDIATRICS
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
The paediatric national data stream Paediatric Personalized Research Network Switzerland (SwissPedHealth) spans across paediatric disciplines and is built in a scalable and modular way in terms of governance, data infrastructure, and patient and public involvement to enable a gradual increase in coverage of healthcare data from the Swiss child population.
SwissPedHealth is part of the Swiss Personalized Health Network and Personalized Health and Related Technologies Swiss personalised health ecosystem and collaborates with paediatric teaching hospitals, focusing on personalised health research through a harmonised dataset across Switzerland and adhering to a standard interoperability framework based on the Resource Description Framework.
Regulatory challenges include complex legal agreements, multicentre ethics proposals and data harmonisation requiring both technical and clinical expertise.
Differences in hospital systems and data availability lead to challenges in creating comparable datasets and necessitating central coordination and additional resources.
In the future, expanding data harmonisation efforts to non-university clinics and primary care, as well as other data sources such as federal data, cohorts or registries will be essential to include a broader segment of the Swiss child population, which can be built on SwissPedHealth in a stepwise approach.
Introduction
In Switzerland, over 85 000 children are born annually, contributing to a current population of 1.7 million inhabitants aged under 20 years.1 Every year, over 100 000 children require admission as inpatients to hospitals. Many more are seen in outpatient clinics. Children represent a vulnerable population, with unique developmental physiology, dependence on adults for care-seeking and disease epidemiology. Adverse effects related to diseases, treatments, environmental exposures and lifestyle during early childhood will affect children themselves, their families and future offspring for many decades, markedly increasing the associated direct healthcare and other indirect costs for society.2–5 Evidence to guide best paediatric healthcare remains sparse. At present, the evidence base for many diagnostic and therapeutic procedures is either absent, of low quality (often data from small cohorts) or inferred from adults. Paediatric evidence generation often suffers from a long lag time from results to implementation. While these features apply to the paediatric population in general, the limitations are aggravated in Switzerland due to the fragmentation of its healthcare system with regional isolated data repositories. In addition, multiple paediatric registries coexist in Switzerland, and they lack interoperability. Overall, the present state of paediatrics thus lacks preparedness to appropriately develop, evaluate or adapt to the rapidly increasing possibilities for intelligent decision-support tools which could enhance the management of paediatric patients.
Over the past decades, progress in social and preventive health and new therapies have reduced the morbidity and mortality caused by common conditions such as infections, cancer or trauma. Consequently, modern paediatrics in high-income countries like Switzerland deals with a conundrum of rare diseases that present as unique and often life-threatening phenotypes and require highly personalised approaches.6–10 In addition, common problems such as obesity-related morbidity and chronic respiratory conditions frequently result in lifelong trajectories reaching out well into adulthood.11 12 Furthermore, although patients and parents have intimate knowledge of the disease and needs, they remain poorly integrated into healthcare planning and research.
The demand to continuously improve paediatric data for research and improved care requires better digitalisation, harmonisation and automated extraction of routine clinical data. This represents a unique opportunity to break silos and to shape a data-driven, agile, learning healthcare system that will deliver better, more efficient and more personalised care to children and their families. SwissPedData, an earlier Swiss initiative, defined an agreed set of common data elements from routine healthcare records to gather standardised and high-quality information on key aspects of interest to clinicians and researchers for paediatric clinical care.13 14 This paediatric core dataset has the potential to be enriched with patient-reported outcomes, population-based and administrative information, specialised disease registries and high-density data from advanced clinical diagnostics and research projects such as omics technologies.
As part of a national call for data streams (multidisciplinary consortia research platforms), we designed the Paediatric Personalized Research Network Switzerland (SwissPedHealth). The main goal of SwissPedHealth is to implement routine clinical data extraction pipelines in paediatric hospitals and make this core dataset available for research, benchmarking and improvement of quality of care, thereby implementing the infrastructure towards a learning national paediatric health system. This study describes the design, structure, aims and current achievements of the national data stream (NDS) SwissPedHealth.
Methods and analysis
SwissPedHealth
SwissPedHealth (https://www.swisspedhealth.ch/) is a paediatric NDS that was started in September 2022 aiming to make health-related personal data from routine clinical practice collected from hospitals in Switzerland interoperable, standardised, quality-controlled and ready for observational research, clinical trials, personalised medicine, health-policy research and clinical audits. Several quality improvement and research projects will test the infrastructure for routine clinical data collection and enrich the data stream.
The SwissPedHealth data stream is embedded in an ecosystem supported by two funding bodies: the Swiss Personalized Health Network (SPHN, https://sphn.ch/) and the Strategic Focal Area ‘Personalized Health and Related Technologies’ (PHRT) of the ETH Domain (Swiss Federal Institutes of Technology, https://www.sfa-phrt.ch/). SPHN and PHRT aim to build a Swiss personalised health ecosystem, improve research and innovation in healthcare by developing industrialised infrastructures for efficient secondary use of electronic health record data across hospitals, research institutions, biobanks and registries in Switzerland and enhance the quality of the collected data.15 The Data Coordination Center (https://sphn.ch/network/data-coordination-center/) is managed by the personalised health informatics group of the Swiss Institute of Bioinformatics (https://www.sib.swiss/). The Center promotes, designs, implements and coordinates efforts around data semantics and standards across Swiss hospitals and other data providers. SIB provides legal support and coordination within the SPHN framework. BioMedIT (https://www.biomedit.ch/) acts as a national secure computing network for health-related data.
The SwissPedHealth consortium includes clinical partners from all five Swiss paediatric university hospitals (Universitäts-Kinderspital beider Basel, Inselspital Bern, Hôpitaux universitaires de Genève, Centre hospitalier universitaire Vaudois and University Children’s Hospital Zurich); two non-academic hospitals (Luzerner Kantonsspital and Ostschweizer Kinderspital); three research institutions: the Federal Institutes of Technology ETH Zurich (Eidgenössische Technische Hochschule), EPFL (École Polytechnique Fédérale de Lausanne), and the Institute of Social and Preventive Medicine (University of Bern); and patient and public representatives. The consortium unites expertise in clinical paediatrics and paediatric research (data science), omics, machine learning (ML), patient and public involvement, and public health. SwissPedHealth builds on previous projects, including SwissPedData13 14 and the Swiss Research Network of Clinical Paediatric Hubs (SwissPedNet, https://www.swisspednet.ch/home).
SwissPedHealth governance structure and regulatory framework
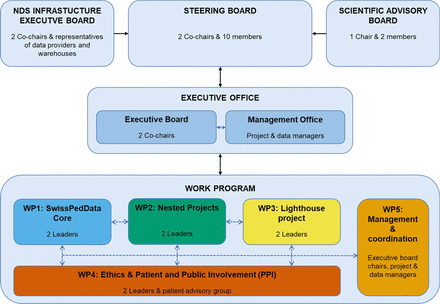
The governance structure of SwissPedHealth consists of a steering level with an overarching Steering Committee, an Infrastructure Executive Board and a Scientific Advisory Board; a management level comprising the Executive Office (Executive Board and Management Office) and an operational level with five work packages covering the data integration and research programmes (figure 1). The regulatory framework of SwissPedHealth builds on existing collaboration agreements through SwissPedNet as well as template documents from SPHN. SwissPedHealth uses a three-tiered structure, namely, Tier 1—SwissPedNet Service Level Agreements, Tier 2—SwissPedHealth Infrastructure Consortium Agreement and Tier 3—project-specific Data Project Consortium Agreements (DPCA). Tier 1 regulates the data flow from participating Children’s Hospital to their clinical data warehouses (CDWs). Tier 2 issues guidance and templates to access clinical data through SwissPedHealth, including General Terms and Conditions for data projects, Data Transfer and Use Agreements, and DPCA templates. Tier 3 focuses on project-specific governance aspects, and details of project-specific DPCA elements including project aims, sponsorship, project partners, data needs and timelines. Principles of data governance and regulatory compliance respect primary ownership of routine data by the hospitals with their respective CDWs and research-only data by the respective research teams. Requests to access data are governed accordingly at the level of the NDS Data Access Request Committee of the NDS Infrastructure Executive Board. The Infrastructure Consortium Agreement has been approved by the seven participating children’s hospitals and all three BioMedIT nodes.
SwissPedHealth governance. NDS, national data stream; SwissPedHealth, Paediatric Personalized Research Network Switzerland; WP, work package.
SwissPedHealth data access regulation
Access to the NDS data is regulated through the SwissPedHealth Infrastructure Consortium Agreement. While planning to carry out a project using data from the SwissPedHealth NDS, researchers will need to set up a DPCA using our template and involving all participating partners. This should include ‘internal’ SwissPedHealth partners and ‘external’ partners, that is, those who are not part of the SwissPedHealth NDS Infrastructure Consortium. For the DPCA, projects should consider the SwissPedHealth General Terms and Conditions and, wherever possible, try to adopt them without or with minor deviations. This will facilitate the process of legal review and signing for all internal partners since the General Terms and Conditions will have already been reviewed as part of the development and adoption process. If deviations are required, they will need to be specified in the DPCA and should be highlighted during its legal review. The following additional documentation will be required to complete the DPCA: project funding application/proof of funding, study protocol and ethical approval. In case the latter is not available it should be added as soon as available, and the DPCA cannot be executed without evidence of ethical review and approval. All DPCAs will need to incorporate a Data Transfer and Use Agreement and a Data Transfer and Processing Agreement. These are included in the SwissPedHealth DPCA template. Projects need to set up a specific space in the BioMedIT secure IT network for data access and processing. Research groups access their B-spaces through the BioMedIT Portal, where they manage users, encryption keys and data transfers, while also accessing support and tools. Researchers use two-factor authentication for secure access and only extract nonsensitive or aggregated results, keeping sensitive data within the platform. Adaptations to the agreements may be necessary in cases where the Data Processors are not one of the BioMedIT nodes (SENSA—Lausanne, SciCORE—Basel and SIS—Zurich). For the Data Transfer and Use Agreement section, each project will need to describe and depict the planned data flow as well as data and metadata to be transferred. Details on the process to access the data and contact information will be made available through the SPHN metadata catalogue, as well as information on data sources and qualitative and quantitative metadata on concept availability in the NDS.
Data management
Data flow and FAIRification
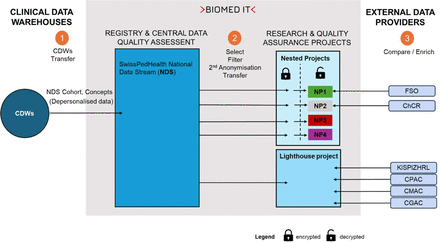
Routine clinical data is extracted from the electronic data capture systems and other source systems and loaded into the CDWs of each children’s hospital from where it can be transferred to the NDS. Data are harmonised according to the SPHN ontology in each CDW. Data that form part of the NDS are planned to be indexed as part of the data FAIRification process to make data Findable, Accessible, Interoperable and Reusable.16 17 These FAIR principles lay the basis of common practices among hospitals and facilitate the further use of routine clinical data for research. The data from the different hospital source systems are transformed and mapped to the SwissPedHealth project-specific schema in the CDWs. The project-specific schema extends the SPHN schema to cover the needs of the SwissPedHealth NDS. To achieve an interoperable data representation, that is coherent and semantically understandable both for humans and machines, SPHN employs concepts that incorporate existing ontologies and terminologies such as Systematic Nomenclature of Medicine—Clinical Terms (SNOMED CT, https://www.snomed.org).15 18 Concepts are based on the Resource Description Framework (RDF, https://www.w3.org/RDF/). CDWs use the SPHN Connector tool (https://git.dcc.sib.swiss/sphn-semantic-framework/sphn-connector) to pseudonymise and de-identify data and to transform it to RDF format compliant with the SPHN schema. RDF data integrity checks and data queries can be done using Shapes Constraint Language and SPARQL, an RDF Query Language. Data are transferred from CDWs to the secure project space (the so-called ‘B-space’) for the SwissPedHealth NDS at the sciCORE BioMedIT node in Basel via the Secure Encryption and Transfer Tool (https://gitlab.com/biomedit/sett) (figure 2). Data transfers from CDWs to the NDS B-space are planned to occur recurrently.
SwissPedHealth data flow. Numbers indicate: 1) Data flow from CDWs to NDS. Data transfer requests triggered by NDS B-space to CDWs, transfer via the Secure Encryption and Transfer Tool (SETT) tool. 2) Data flow from NDS to project specific spaces. Data transfer triggered by nested projects B-space or lighthouse B-space to NDS B-space. 3) Data flow from external data providers to project specific spaces. CDW, clinical data warehouses; CGAC, Clinical Genomic Analysis Centre; ChCR, Swiss childhood cancer registry; CMAC, Metabolomics Analysis Centre; CPAC, Clinical Proteomics Analysis Centre; FSO, Federal Statistical Office; KISPIZHRL, University Children’s Hospital (Kispi) Zurich Research Lab; LH, Lighthouse project; NDS, National Data Stream; NP, nested project; NP1, SwissPedGrowth; NP2, SwissPedCancer; NP3, SwissPedLung; NP4, SwissPedAntibio; SwissPedHealth, Paediatric Personalized Research Network Switzerland.
The NDS B-space serves as a data repository, where the central data managers perform data quality assessment and remediation as needed, data filtering and transfer to separate project-specific B-spaces for research or quality improvement projects (‘workspaces’). Currently, SwissPedHealth holds two separate workspaces for data analyses: one for the nested projects and the other for the lighthouse project. The access to these separate project-specific workspaces is coordinated by permission managers and restricted to permitted users. Users must perform the mandatory data protection and IT security awareness training of SPHN and accept the sciCORE Terms of Use. Secure and encrypted modalities provided by the SPHN infrastructure are used for all data transfers between CDWs and BioMedIT, and between external data providers and BioMedIT.
Data quality assessment
The central data quality assessment includes the evaluation of data validity, accuracy, completeness, consistency, timeliness and integrity on the NDS B-space, both individually for each CDW and across CDWs to investigate potential discrepancies between data providers. The output of this report will be fed back to the CDWs for clarification and checks as necessary to iteratively improve data quality on the NDS B-space. Data will be transferred together with a data quality report to the project-specific workspaces, where researchers will perform additional data quality assessments tailored to project needs. In particular, one of the nested projects is specifically designed to assess data quality completeness, representativeness and accuracy by comparing the datasets for children diagnosed with cancer against an external reference standard: the national Childhood Cancer Registry (ChCR).
Nested projects
SwissPedHealth includes four nested projects that serve as use cases to address relevant questions in child health, focusing on (1) anthropometrics—SwissPedGrowth, (2) childhood cancer—SwissPedCancer, (3) paediatric lung function and respiratory diseases—SwissPedLung and (4) antibiotic utilisation—SwissPedAntibio (table 1). These projects showcase the feasibility and usefulness of the NDS, using selected key concepts from the SwissPedData core dataset from a large number of children admitted to Swiss hospitals or visiting outpatient clinics, and at the same time serve as data quality assurance.19 20 Health-related personal data obtained in clinical routine, such as demographic and treatment data as well as medical history captured in electronic health records of patients from participating children’s hospitals, will be extracted for the years 2017–2023. In the future, regular ongoing data transfers are planned. Transferred data will include SPHN concepts (https://www.biomedit.ch/rdf/sphn-schema/sphn/2024/1), such as diagnostic codes (SNOMED CT, 10th revision of the International Classification of Diseases (ICD-10), International Classification of Diseases for Oncology, 3rd Edition (ICD-O3)), age, height and weight, Logical Observation Identifiers Names and Codes for lab results, and Anatomical Therapeutic Chemical Classification System codes for drugs.
Specifications of the four nested projects within SwissPedHealth: SwissPedGrowth, SwissPedCancer, SwissPedLung and SwissPedAntibio
All nested projects test the availability and quality of information from the electronic healthcare records, pilot procedures and data flows. The results of the nested projects will feed back to hospital information systems, and national registries if applicable, to progressively improve data quality. The nested projects are tasked to explore and define strategies to link patient data (eg, socioeconomic, nationality and birth record data) with the Swiss National Cohort21 22 and the Federal Statistical Office and compare datasets with external reference standards such as the Swiss ChCR (https://www.childhoodcancerregistry.ch/).23 24 If successful, these linkage strategies will enhance the scalability and interconnectedness of SwissPedHealth, and allow additional assessment of data quality, such as consistency, accuracy and completeness. Furthermore, the nested projects will partner with international research networks to identify best practices, compare performance across countries and enhance the learning of paediatric health systems. Full detailed study protocols for the nested projects, including specific data analysis plans, will be published separately.
Lighthouse project: development of a multi-omics workflow to discover rare diseases in children with life-threatening phenotypes
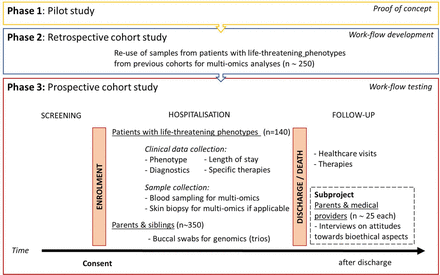
The SwissPedHealth lighthouse project focuses on improving the diagnosis of rare diseases in children with potentially life-threatening conditions by integrating clinical data with multi-omics data using ML algorithms. This project will showcase how the NDS can be enriched with biological data for a specific cohort, in this case children with suspected rare diseases. Rare diseases predominantly affect children, frequently causing premature death or chronic disability.25 The discovery of rare diseases has been rapidly increasing in past decades due to decreasing cost coupled with the rising speed of genome sequencing.26–28 However, even recent rapid whole-exome or whole-genome sequencing programmes in highly selected patients report a diagnostic yield of only 30–50%,29 and the challenge has shifted from identifying genetic alterations to defining their functional relevance. Multilayered omics technologies define relationships between genes, proteins, metabolites and phenotypic traits to accelerate identification of the underlying cause of rare clinical presentations.30 The lighthouse project will progress through three phases. It builds on our previous pilot testing multi-omics in a defined metabolic disease (phase 1)31 to first develop (phase 2) and then prospectively assess (phase 3) a multi-omics workflow designed to identify rare diseases in children with life-threatening conditions (figure 3).
Study design of the SwissPedHealth lighthouse project. Development of a multi-omics workflow to discover rare diseases in children with life-threatening phenotypes. SwissPedHealth, Paediatric Personalized Research Network Switzerland.
Phase 1 provided a proof of concept for integrative multi-omics analyses in the inborn error of metabolism—methylmalonic aciduria.31 In phase 2, we access existing cohorts of children with different life-threatening phenotypes (target n=250), including those who presented with homocystinuria and have a suspected underlying genetic basis, the Swiss Paediatric Sepsis Study,32 the Genetic Study of Immunodeficiency (GSI)33, the Childhood Life-Threatening Infectious Disease Study34 and the Kennedy fellowship cohort.35 We apply an integrated approach of whole-genome sequencing, RNA sequencing, proteomics36–40 and metabolomics to identify rare deleterious gene variants and demonstrate their impact at the gene, transcript and proteome/metabolome level. Samples are processed by the Clinical Genome Analysis Centre, the Clinical Proteotype Analysis Centre and the Clinical Metabolomics Analysis Centre. These centres are all part of the Swiss Multi-Omics Centre (http://smoc.ethz.ch/), which is integrated into the PHRT secure computational infrastructure and provides secure data processing and sharing to BioMedIT. Through the lighthouse project, we seek to develop novel bioinformatic pipelines and computational methods using ML to integrate multi-omics data with clinical information. This multi-omics workflow will create a diagnostic tool that can be optimised for rapid result generation. In phase 3, we will test and evaluate the workflow developed in phase 2 using a cohort of children prospectively recruited from partner Swiss hospitals (target n=140). Children with potentially life-threatening conditions where a rare disease is suspected are eligible. Screening occurs through healthcare staff in the intensive care units and specialist departments as well as by research staff. The eligibility of a patient is assessed by a panel of consortium members, including experts in genetics, metabolism, neurology, immunology or intensive care, before proceeding to seeking informed consent. For each enrolled patient, we collect blood samples (PAX gene for RNA extraction, ethylene-diamine-tetraacetic acid for DNA extraction and serum for proteome/metabolome assessment), and, where feasible, skin biopsies (fibroblasts). In addition, we seek consent to obtain mucosal swabs from parents and siblings of included patients for DNA extraction for trio analyses. We also obtain consent for the encrypted use of patients’ and relatives’ (genetic) data and biological material for future research. Samples are preprocessed at hospitals and sent in batches to the Swiss Multi-Omics Centre for multi-omics data generation. Recruitment for phase 3 was started in September 2022 at the University Children’s Hospital Zurich and will be expanded to other participating children’s hospitals.
Analysis outline for the lighthouse project
Overall, the aim is to comprehensively assess the impact of genetic determinants on disease and their relationships with RNA, protein and other metabolites. To search for novel disease-causing variants both in phases 2 and 3, we will use an optimised workflow for automated DNA variant calling and annotation, providing ready-to-interpret reports. Rare variants will be prioritised based on (1) the potentially damaging effect of the variant, (2) the possible known function of the corresponding gene in the clinical presentation and (3) the degree of purifying selection to which they are subjected. RNA-seq data analysis will be used for quantitative expression profiling and alternative splicing analysis. DNA variants, RNA transcripts, proteoforms, and metabolites will be mapped onto biological networks and pathways using tools.
A joint analysis will include statistical genomics to detect the enrichment of disease-causing variants within specific phenotypes. To investigate genotype-phenotype relationships, we will examine genetic determinants of disease and their impact on RNA, protein and metabolite levels. To analyse rare (or combined rare and common) DNA variation, we will use the same methods as common variant analysis with the addition of several necessary protocols. These methods will be used to analyse both rare and common DNA variants simultaneously, and rare variants exclusively, while adjusting for relevant covariates and incorporating multivariate and multicategory outcome models. Furthermore, we will perform gene- and protein-based pathway collapse analysis, incorporating covariates, to summarise variant-level information within functional units or biological pathways.
We will perform RNA-seq data analysis using regression models to assess the relationship between gene expression and phenotypic outcomes while adjusting for potential confounders. For proteomic and metabolite analysis, we will map proteoforms and metabolites onto biological networks and pathways using enrichment analysis methods and apply multivariate statistical methods to identify patterns and relationships between proteomic or metabolomic profiles and phenotypes. Throughout the study, the major outcome variables will consist of extreme phenotypes such as inborn errors of metabolism and inborn errors of immunity. Strict thresholds for multiple test correction will be applied based on the number of tests. Candidate causal determinants will be interpreted based on best practices for clinical genetic reporting and American College of Medical Genetics and Genomics recommendations.41 42
We additionally leverage ML for the analysis of the multi-omics data. In the context of this study, patients are characterised by means of multiple representations, or views—clinical, genomic, transcriptomic, proteomic and metabolomic—each critically relevant to the development of a rare disease. While previous studies mostly used self-supervised approaches such as autoencoders,43 we will explore multimodal and multitask learning to find an optimal subset of explanatory genes and aim to develop novel ML-based approaches to the analysis of multi-omics datasets.44–49
Patient and public involvement and engagement and Bioethics
Patient and public involvement and engagement (PPIE) represents an integral part of research conduct to increase the quality, meaningfulness, credibility and impact of clinical research. Building on existing patient representatives and PPIE groups, SwissPedHealth engages with a Patient and Family Advisory Group with diverse members from French and German speaking regions in Switzerland with different roles and contributions (figure 4). In addition to parents, we will include both adolescent patients and adult survivors of childhood illnesses. The Patient and Family Advisory Group is chaired by two PPIE representatives who are part of the Steering Committee. Together with PPIE representatives, we develop and test a PPIE toolbox to guide future paediatric studies in Switzerland. This toolbox will review PPIE involvement and propose practical guidelines that can be used at different time points during the design and conduct of research, as well as the dissemination of study results.
Patient and public involvement activities in SwissPedHealth. SwissPedHealth, Paediatric Personalized Research Network Switzerland.
Bioethical aspects surrounding perceptions on omics analyses will be explored in depth within a sub-project of the lighthouse project. Given the future goal to translate the current research multi-omics workflow into a clinical standard available to healthcare providers for patients with suspected genetic rare diseases, it is important to ascertain how this information is viewed by end-users, such as clinicians, patients and family members. We will interrogate the perspectives of up to 25 medical providers and 25 parents whose children participate in phase 2 of the lighthouse project by conducting exploratory interviews to determine their attitudes towards a hypothetical offer and return of results and other aspects that will help inform future clinical translation of these technologies.
Use and productivity assessment plan for SwissPedHealth
To assess the use of our data infrastructure, key metrics include the number of projects requesting data access and establishing regulatory documents, the volume of data requests for approved projects in BioMedIT workspaces, users per workspace and data access frequency. Support requests from project-specific workspaces are analysed to identify and evaluate potential bottlenecks. The productivity of SwissPedHealth is assessed by monitoring research publications, citations, conference presentations, new collaborations, project plans and grants. PPIE activity is recorded through minutes of focus groups, satisfaction surveys, number of projects and researchers using the planned PPIE toolbox and tracking outreach activities.
Ethics and dissemination
Ethical approvals
The SwissPedHealth database regulations SwissPedHealth was approved based on the advice of the ethical committee Northwest and Central Switzerland (EKNZ, AO_2022-00018). These regulations have been drawn up in compliance with the applicable standards, considering the Federal Law on Research Involving Human Subjects (Art. 51 Human Research Act), cantonal legislation and the Federal Data Protection Act. Each nested research or quality improvement project conducted under SwissPedHealth is subject to standard ethical regulatory requirements. Separate ethical approvals or waivers for each nested research were obtained: SwissPedGrowth (ethical committee (EC) Bern, b2023-00022), SwissPedCancer (EC Bern, Req-2023-01081), SwissPedLung (EC Bern, under review), SwissPedAntibio (EKNZ, Req-2023-01501), the lighthouse project (EC Zurich, 2022-00351) and the related bioethics interview project (EC ETH Zurich, 2023-N-253).
Dissemination
The findings of SwissPedHealth, including the lighthouse and nested projects, will be published in peer-reviewed journals and scientific conferences, as well as disseminated to patients, families and the public. Through dedicated PPIE focus groups, we will design dissemination to other media, such as the press, radio, television, podcasts and social media, to increase awareness about the NDS and to promote its use for future research. We will work with families to display understandable and relevant information on our website (https://www.swisspedhealth.ch/).
SwissPedHealth will promote access to analysable datasets and open-access publications. Research outcomes will be made publicly available according to the FAIR principles and in line with the policies and regulations of the partner institutions and guided by the SPHN and PHRT principles.
Discussion
The paediatric NDS SwissPedHealth digitalises and harmonises the use of routine clinical data for clinical research and evidence generation. It will revolutionise paediatric healthcare in Switzerland in several ways: (1) personalised care: by harnessing patient data through tailoring healthcare to individual children’s needs, ensuring the best possible outcomes; (2) benchmarking: by enabling hospitals to compare their performance with others, identifying areas for improvement; (3) informing policy decisions: policymakers will have access to data-driven insights, aiding in the development of effective healthcare policies and (4) enabling clinical and therapeutic studies: researchers will be able to use the NDS data to design and conduct studies that lead to more effective treatments for children. SwissPedHealth strives to set up a sustainable framework and infrastructure to conduct Swiss-wide observational and interventional studies, enhancing clinical trial readiness, and to develop a learning healthcare system driving frontier, innovative and highly patient-focused research. It seeks to integrate clinical, federal administrative data and research data across leading Swiss hospitals under joint governance. SwissPedHealth will thereby overcome the challenge of paediatric data being kept in local silos, in nonstandardised and non-interoperable forms, highly dispersed across hospitals, and therefore not usable for translational research and clinical trials of sufficient power.
Strengths and limitations
SwissPedHealth is embedded in the SPHN and PHRT Swiss personalised health ecosystem and builds on existing and endurable partnerships with all paediatric teaching hospitals centred around SwissPedNet. It will implement a harmonised paediatric health dataset across Switzerland, which is scalable to additional datasets and sites, and serves as a high-visibility example of how this infrastructure can be enriched with high-density data to deliver cutting-edge research on personalised health. SwissPedHealth also builds on previous successful national multicentre initiatives such as SwissPedData, the Swiss Pharmacokinetics Clinical Data Warehouse project (SwissPKcdw), the Swiss Paediatric Sepsis Study and the GSI. The broad scope of SwissPedHealth also favours potential synergies with other current SPHN- and PHRT-funded projects (https://sphn.ch/network/projects/, such as LUCID, IICU, SPHN-SPAC, SwissPedDW, SwissPedHealth-PREPP, ADONIS and INFRA). Internationally, SwissPedHealth shares the goals and strategies of several initiatives, such as the European Children’s Hospital Organization (https://www.echohospitals.org/), and the US-based paediatric clinical research network (https://pedsnet.org/) which use a common interoperable data platform to optimise the use of electronic health records.
There are several challenges to be overcome in SwissPedHealth. Regulatory processes, such as legal agreements and multicentre ethic proposals, that need to be signed by several hospitals and institutions are cumbersome, despite standardised templates. The design of RDF concepts requires multidisciplinary expertise in content aspects, like medicine and biology, as well as RDF, semantic and technical knowledge. Data harmonisation efforts at local CDWs are time-consuming and require both technically and clinically skilled staff and adequate local resources. Source systems are different in every hospital, making efforts in one site often not applicable or directly comparable to other sites. Central coordination efforts are crucial to create synergies whenever possible. There is heterogeneity in data availability between CDWs. Thus, data gaps are expected for the nested and lighthouse projects of SwissPedHealth. Regulatory barriers need to be overcome to integrate patient data from sources other than hospitals such as registries and federal offices which have their own regulations and data structure. Acceptance rates of General Consent or Informed Consents and regulatory allowances will affect the representativeness of the study populations for projects conducted with SwissPedHealth. Furthermore, data harmonisation efforts have primarily focused on university hospitals; however, additional efforts are necessary to extend coverage to non-university clinics and primary care, thereby including a broader segment of the Swiss child population. Future endeavours will facilitate and explore data interoperability with existing international data models such as Observational Medical Outcomes Partnership Common Data Model.15
Outlook
We will set up a sustainable framework and infrastructure to conduct Swiss-wide observational and interventional studies, enhancing clinical trial readiness, and to develop a learning healthcare system driving frontier, innovative and highly patient-focused research. In the future, the experience gained and the infrastructure and procedures developed within SwissPedHealth are expected to facilitate the integration of other Swiss paediatric hospitals and linkage with different databases, such as Swiss paediatric national registries, as well as exchange with international consortia.
The vision of SwissPedHealth is a future where routine paediatric data from hospitals, and ideally also primary care providers, other research institutions, biobanks and registries, can be securely used to sustain a learning healthcare system and for research to improve and personalise healthcare of children. This framework within a Swiss personalised health ecosystem is only possible through joint, and highly coordinated efforts from clinicians, engineers, scientists, researchers, legal and regulatory experts, and patients and families.
Ethics statements
Patient consent for publication
Acknowledgments
We would like to thank all the patients and families who have been providing advice on SwissPedHealth and its projects, as well as the clinical and research teams at the participating institutions.
References
Footnotes
RM and FNB are joint first authors.
JEV, LJS, JAB and CEK are joint senior authors.
X @effyvayena
Collaborators SwissPedHealth consortium: Andrea Agostini (Department of Computer Science, Institute for Machine Learning, ETH Zurich, Zurich, Switzerland), Anita Rauch (Institute of Medical Genetics, University of Zurich, Zurich, Switzerland), Anna Hartung (Inselspital, Bern University Hospital, University of Bern, Switzerland), Audrey van Drogen (PHRT Swiss Multi-Omics Centre [SMOC], ETH Zurich, Zurich, Switzerland & Institute of Translational Medicine [ITM], Department of Health Sciences and Technology [D-HEST], ETH Zurich, Zurich, Switzerland), Aurélie Martin Necker (Patient and Family Advisory Committee, SwissPedHealth), Ben D Spycher (Institute of Social and Preventive Medicine [ISPM], University of Bern, Bern, Switzerland), Christian Kahlert (Ostschweizer Kinderspital, St Gallen, Switzerland), Christopher B Forrest (Center for Applied Clinical Research, Children’s Hospital of Philadelphia, Philadelphia, USA), Claudia E Kuehni (Institute of Social and Preventive Medicine [ISPM], University of Bern, Bern, Switzerland & Division of Paediatric Respiratory Medicine and Allergology, Children's University Hospital, Inselspital, University of Bern, Bern, Switzerland), Cornelia Hagman (Department of Intensive Care and Neonatology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), D Sean Froese (Division of Metabolism and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Daphné Chopard (Department of Computer Science, Institute for Machine Learning, ETH Zurich, Zurich, Switzerland & Department of Intensive Care and Neonatology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Dylan Lawless (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland & Department of Intensive Care and Neonatology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Effy Vayena (Department of Health Sciences and Technology, Institute of Translational Medicine, ETH Zurich, Zurich, Switzerland), Eirini I Petrou (Department of Health Sciences and Technology, Institute of Translational Medicine, ETH Zurich, Zurich, Switzerland), Emanuele Palumbo (Department of Computer Science, Institute for Machine Learning, ETH Zurich, Zurich, Switzerland), Eric Giannoni (Clinic of Neonatology, Lausanne University Hospital, University of Lausanne, Lausanne, Switzerland), Fabiën N Belle (Institute of Social and Preventive Medicine [ISPM], University of Bern, Bern, Switzerland), Ioannis Xenarios (PHRT Swiss Multi-Omics Centre [SMOC], EPFL, Lausanne, Switzerland & Department of Computational Biology, University of Lausanne, Lausanne, Switzerland & Health 2030 Genome Center, Foundation Campus Biotech, Geneva, Switzerland), Jacques Fellay (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland & Biomedical Data Science Center, Lausanne University Hospital, University of Lausanne, Lausanne, Switzerland), Jana Pachlopnik Schmid (Division of Immunology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Julia A Bielicki (Paediatric Research Center, University Children's Hospital Basel [UKBB], University of Basel, Basel, Switzerland & Centre for Neonatal and Paediatric Infection, St George’s, University of London, London, UK), Julia E Vogt (Department of Computer Science, Institute for Machine Learning, ETH Zurich, Zurich, Switzerland), Kathrin Hofmann (Patient and Family Advisory Committee, SwissPedHealth), Katrin Männik (PHRT Swiss Multi-Omics Centre [SMOC], EPFL, Lausanne, Switzerland & Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland & Health 2030 Genome Center, Foundation Campus Biotech, Geneva, Switzerland), Keith Harshman (PHRT Swiss Multi-Omics Centre [SMOC], EPFL, Lausanne, Switzerland & Health 2030 Genome Center, Foundation Campus Biotech, Geneva, Switzerland), Kelly Ormond (Department of Health Sciences and Technology, Institute of Translational Medicine, ETH Zurich, Zurich, Switzerland & Department of Genetics, Stanford University School of Medicine, Stanford, California, USA), Klara Posfay-Barbe (Hôpitaux Universitaires de Genève, Geneva, Switzerland), Léa Ho Dac (Division of Paediatric Respiratory Medicine and Allergology, Department of Paediatrics, Inselspital, Bern University Hospital, University of Bern, Switzerland), Lorenz M Leuenberger (Institute of Social and Preventive Medicine [ISPM], University of Bern, Bern, Switzerland), Luregn J Schlapbach (Department of Intensive Care and Neonatology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland & Child Health Research Centre, The University of Queensland, Brisbane, Australia), Manon Jaboyedoff (Pediatric Infectious Diseases and Vaccinology Unit, Service of Pediatrics, Department Mother-Woman-Child, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland), Mariam Ait Oumelloul (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), Martin Stocker (Luzerner Kantonsspital, Luzern, Switzerland), Matthias R Baumgartner (Division of Metabolism and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Nicola Zamboni (PHRT Swiss Multi-Omics Centre [SMOC], ETH Zurich, Zurich & Institute of Molecular Systems Biology, ETH Zurich, Zurich, Switzerland), Nicole Goebel (Research and Analyses Services, Digitalisation & ICT Division, University Hospital Basel, Basel, Switzerland), Patrick G A Pedrioli (PHRT Swiss Multi-Omics Centre [SMOC], ETH Zurich, Zurich, Switzerland & Institute of Translational Medicine [ITM], Department of Health Sciences and Technology [D-HEST], ETH Zurich, Zurich, Switzerland & Swiss Institute of Bioinformatics, Lausanne, Switzerland & Department of Biology, Institute of Molecular Systems Biology, Swiss Federal Institute of Technology/ETH Zürich, Zurich, Switzerland), Philipp Latzin (Division of Paediatric Respiratory Medicine and Allergology, Department of Paediatrics, Inselspital, Bern University Hospital, University of Bern, Switzerland), Rebeca Mozun (Department of Intensive Care and Neonatology and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Roger Lauener (Ostschweizer Kinderspital, St Gallen, Switzerland), Sandra Goetze (PHRT Swiss Multi-Omics Centre [SMOC], ETH Zurich, Zurich, Switzerland & Institute of Translational Medicine [ITM], Department of Health Sciences and Technology [D-HEST], ETH Zurich, Zurich, Switzerland), Seraina Prader (Division of Immunology and Children’s Research Centre, University Children’s Hospital Zurich), Simon Boutry (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland), Sven Schulzke (Department of Neonatology, University Children's Hospital Basel [UKBB], University of Basel, Basel, Switzerland), Tatjana Welzel (Paediatric Research Center, University Children's Hospital Basel [UKBB], University of Basel, Basel, Switzerland), Thomas M Sutter (Department of Computer Science, Institute for Machine Learning, ETH Zurich, Zurich, Switzerland), Varvara Dimopoulou (Clinic of Neonatology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland), Vito RT Zanotelli (Division of Metabolism and Children’s Research Centre, University Children’s Hospital Zurich, University of Zurich, Zurich, Switzerland), Xeni Deligianni (Research and Analyses Services, Digitalisation & ICT Division, University Hospital Basel, Basel, Switzerland), Xenia Bovermann (Division of Paediatric Respiratory Medicine and Allergology, Department of Paediatrics, Inselspital, Bern University Hospital, University of Bern, Switzerland), Yara Shoman (Institute of Social and Preventive Medicine [ISPM], University of Bern, Bern, Switzerland).
Contributors RM, FNB, JF, CBF, DSF, EG, SG, PL, RL, KO, JPS, PGAP, KMP-B, AR, SMS, MS, BDS, EV, NZ, JEV, LJS, JAB and CEK designed the work and applied for funding. KH and AMN contributed as patient representatives. RM, FNB, TW and AA contributed to the project conduct, project management and data management. RM and FNB drafted the manuscript. All authors reviewed and approved the manuscript. LJS acted as guarantor. Members of the SwissPedHealth consortium not listed above include PhDs, postdocs, scientists and researchers who are involved in the acquisition, processing, analyses or interpretation of the data. JEV, LJS, JAB and CEK share last authorship.
Funding This project was supported through the grant NDS-2021-911 (SwissPedHealth) from the Swiss Personalized Health Network and the Strategic Focal Area 'Personalized Health and Related Technologies' of the ETH Domain (Swiss Federal Institutes of Technology).
Competing interests None declared.
Patient and public involvement Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review Not commissioned; externally peer reviewed.