Article Text
Abstract
Introduction Breaking the chain of transmission of an infectious disease pathogen is a major public health priority. The challenges of understanding, describing and predicting the transmission dynamics of infections have led to a wide range of mathematical, statistical and biological research problems. Advances in diagnostic laboratory procedures with the ability to test multiple pathogens simultaneously mean that co-infections are increasingly being detected, yet little is known about the impact of co-infections in shaping the course of an infection, infectivity, and pathogen replication rate. This is particularly true of the apparent synergistic effects of viral and bacterial co-infections, which present the greatest threats to public health because of their lethal nature and complex dynamics. This systematic review protocol is the foundation of a critical review of co-infection modelling and an assessment of the key features of the models.
Methods and analysis MEDLINE through PubMed, Web of Science, medRxiv and Scopus will be systematically searched between 1 December 2024 and 31 January 2025 for studies published between January 1980 and December 2024. Three reviewers will screen articles independently for eligibility, and quality assessment will be performed using the TRACE (TRAnsparent and Comprehensive Ecological) standard modelling guide. Data will be extracted using an Excel template in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis standard reporting guidelines. This systematic review will apply the SWiM (Synthesis Without Meta-analysis) approach in its narrative synthesis coupled with tables and figures to present data. The synthesis will highlight key dynamical co-infection model features such as assumptions, data fitting and estimation methods, validation and sensitivity analyses, optimal control analyses, and the impact of co-infections.
Ethics and dissemination Ethics approval is not required for a systematic review since it will be based on published work. The output of this study will be submitted for publication in a peer-reviewed journal.
PROSPERO registration number CRD42023481247.
- Health policy
- Protocols & guidelines
- Infection control
- Epidemiology
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
This systematic review will apply the TRAnsparent and Comprehensive Ecological model evaluation approach with a standardised ecological modelling guide.
A comprehensive analysis of research findings, formulation complexities and developments in co-infection modelling techniques will aim to address critical gaps in the existing literature.
To the best of our knowledge, this review will be the first to explore viral-bacterial co-infection epidemic modelling formulations, their underlying assumptions and transmission peculiarities.
This systematic review considers only studies published between January 1980 and December 2024, which may hamper generalisability.
This systematic review investigates only mechanistic mathematical co-infection epidemic modelling approaches, which may create bias.
Introduction
Co-infection refers to the simultaneous infection of a host by two or more pathogens,1 which can include multiple strains of the same infection in a host, as seen with influenza viruses,2–4 or infections by different pathogens within the same host or between different hosts.5–8 Over the years, vaccination campaigns and the use of antibiotics have significantly prevented deaths and helped to combat the transmission of viral-bacterial infectious diseases. However, viral and bacterial infections remain a major public health threat since they can be lethal and predispose the most vulnerable population to further infections.9 For instance, childhood viral-bacterial diseases like pneumonia and diarrhoeal infections remain the main causes of mortality in infants and children, while SARS-CoV-2, HIV, tuberculosis (TB), influenza and hepatitis are among the major lethal viral-bacterial infections in adults globally. According to the WHO 2004 report, viral and bacterial infections of the respiratory system lead with a global burden of 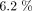 followed by diarrhoeal infections, which account for a burden of
followed by diarrhoeal infections, which account for a burden of  9 The synergism of viral-bacterial co-infections has remained lethal, leading to extensive theoretical and clinical research on the mechanisms behind their transmission dynamics.10 The ecological epidemiology of infectious pathogen interactions and coexistence in the host species can be complex, and their combined effects on disease transmission, clinical outcomes and public health intervention measures remain poorly understood.11–13
9 The synergism of viral-bacterial co-infections has remained lethal, leading to extensive theoretical and clinical research on the mechanisms behind their transmission dynamics.10 The ecological epidemiology of infectious pathogen interactions and coexistence in the host species can be complex, and their combined effects on disease transmission, clinical outcomes and public health intervention measures remain poorly understood.11–13
Many human and animal populations acquire infections with either bacterial or viral pathogens.14 15 Studies on the 2009 pandemic Influenza A (H1N1) in the USA found that within 6 days of influenza infection, a bacterial co-infection occurred. The critically ill patients who developed bacterial co-infections faced an increased risk of death.16 Similarly, epidemiological studies have reported influenza (viral) and Streptococcus pneumoniae (bacterial) co-infections occurring with influenza during influenza seasons, potentially weakening the immunity of individuals which predisposes them to bacteria like S. pneumoniae infections.14 HIV and TB co-infections are common, with individuals infected with HIV often experiencing advanced immunosuppression. This weakened immune response increases their susceptibility to catching a TB infection.17 Therefore, viral-bacterial co-infections are of great public health concern due to the risk factors they present in altering the severity, incidences and mortalities.14
Despite co-infection models having vital implications in public health policy, epidemiology and pathogen evolution, few studies have addressed their complexity and implications in forecasting disease dynamics. Limited systematic reviews have focused on co-infection mathematical epidemiological modelling design, quality and applications. Recent systematic reviews have assessed the impact of co-infections in disease dynamics and have focused on specific co-infections such as HIV-hepatitis, HIV-TB and Ebola-malaria co-infections.11 12 14–20 Some significant and interesting reviews on the impact of co-infections have focused on respiratory tract infections,10 16 aquatic animals21 and modelling within-host co-infections.22
Given the vital significance and potential for wide applications of co-infection studies in public health, understanding the dynamics of the simultaneous coexistence of infectious pathogens is crucial for improving the practicability of co-infection modelling.23 Transition and interaction between classes in a co-infection model may be complex,22 especially in cases where infections present multiple strains or pathogens in a host. Dynamical co-infection epidemiological models are effective tools for highlighting a more comprehensive understanding of infectious disease spread, precise prediction and transition dynamics.24 The models can highlight causal factors, the expected size and duration of an epidemic outbreak, and the extent of resource strain and disease case fatalities associated with the outbreak.25 It is expected of a mathematical model to provide realistic predictions for disease dynamics.24
This systematic review aims to provide an extensive exposition and diagnostics of existing bacterial-viral co-infection epidemiological models that were published from January 1980 to December 2024. The study duration was chosen after reviewing the historical emergence of significant viral and bacterial infections known to cause co-infections, for instance, the HIV/AIDS epidemic (1980s), the SARS-CoV-2 (2002) and the H1N1 influenza (2009) pandemic, which were pivotal moments in infectious disease history that may have influenced co-infection studies. We considered a time frame from January 1980 to December 2024, a duration of 44 years that may give a comprehensive view of the evolution of co-infection studies and capture trends over time. The studies allow us to capture both historical and recent co-infection studies like HIV and tuberculosis, influenza and pneumonia, or COVID-19 and secondary bacterial infections, among others. We will focus on the co-infection model formulation, assumptions, analysis, modelling techniques and data integration while assessing the significance and potential research gaps which might be of interest to mathematical epidemiologists and public health policymakers. The co-infection modelling framework has the potential to guide policies on management and control strategies for emerging and re-emerging infectious diseases.
Research objectives and questions
In the planned systematic review, we will explore and synthesise the mathematical epidemiological modelling of viral-bacterial co-infections and the role of pathogen interactions in shaping transmission dynamics, severity and control of infectious diseases. Its focus is primarily on modelling techniques (assumptions, modelling designs, data fitting and model analysis and control strategies). It will identify research gaps in co-infection modelling mechanisms that may help advance the models to further realism and enhance effective description and forecasting of disease dynamics. Specifically, this systematic review is aimed at addressing the following questions:
How are parameters interrelated in a co-infection model? Does catching a viral pathogen increase or decrease the risk of acquiring a bacterial infection, or is it insignificant and vice versa? Do viral/bacterial pathogen co-infections lead to a higher risk of disease severity?
Does a co-infection alter infectiousness and mortality rates? How are co-infections and mortality rates epidemiologically modelled?
What is (are) the impact(s) of co-infections in shaping the infectious disease spread dynamics?
What are the mathematical modelling approaches applied to describe the dynamics? What research gaps exist in the modelling approaches?
How are data integrated in a co-infection model? What reasonable data inputs/assumptions might be necessary to predict or forecast co-infection disease dynamics? What are the challenges in parameter estimation of a co-infection model?
How can co-infection mathematical models be robustly formulated, analysed, tested and evaluated in a manner that is epidemiologically meaningful and relevant for the provision of guidance on urgent response policy and mitigation of an infectious disease outbreak?
Rationale
The dynamics of pathogen spread in humans and animals can be influenced by co-infections, which may ultimately lead to insignificant, beneficial or detrimental health outcomes.11 The interactions between two or more infectious disease pathogens in host species can have serious outcomes, particularly in vulnerable immunosuppressed individuals such as HIV-positive patients who are at a higher risk of experiencing increased severity or dying due to co-infections.11
Co-infection models offer a way to understand and describe infectious disease spread dynamics and provide both short-term and long-term forecasts. This is done through the integration of biological features and behavioural and environmental epidemiological factors that shape the burden, transmission, surveillance, prevention and control strategies. The last decade has witnessed an increased interest in epidemiological modelling due to its potential to integrate both ecological epidemiology and mathematics under a data, knowledge and strategic decisions framework to explain the transmission and control strategies of infectious diseases.26
Due to the complex nature of the interaction of pathogens for co-infection models, the theoretical design remains a fundamental and pivotal issue that shapes the reliability of estimated parameters, the practicability of intervention measures and realistic forecasting,27 particularly applying methods that account for the scarcity of data which may hamper the biological basis and justifiability of assumptions, integration with data, model analysis and their impact in informing policies in public health.27 28 There is a need to review key aspects of co-infection modelling to understand the milestones achieved, their significance and research gaps and opportunities in co-infection model development that may improve their framework and relevance in informing policy on control interventions and improved health outcomes.
Methods and analysis
Study protocol design
This protocol is reported in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015 checklist.29 30 The review has been registered in PROSPERO.
Eligibility criteria
The following inclusion criteria will apply: (1) full-text research articles written in English on infectious disease mathematical epidemiology, published from January 1980 to December 2024. The choice of English as the language for this review was primarily influenced by the research team’s proficiency in English as well as resource constraints such as time and funding that language translation may demand. A study will be selected if it is investigating viral-bacterial co-infections of infectious diseases using mathematical epidemiological modelling approaches such as deterministic, stochastic and fuzzy logic modelling. (2) Studies focusing on within-host, between-host and multiple-strain co-infections. The exclusion criteria will include (1) articles focused on mono-infection mathematical modelling, clinical experiments and surveys and case studies and (2) duplicate studies, books and book chapters, review protocols and systematic reviews.
Studies search criteria
This systematic review will use the outline for a search strategy provided in Peer Review of Electronic Search Strategies (PRESS) peer review guidelines.31 The search for articles from databases (PubMed, Web of Science, Scopus and medRxiv), selection of studies, data extractions and synthesis will be done comprehensively from December 2024 to April 2025 to identify epidemiological co-infection model studies from January 1980 to December 2024. The search items will be combined via the Boolean operations ‘AND’ and ‘OR’ coupled with words and/or synonyms formed by a combination of statements in four main categories as follows: mathematical approach or term, co-infection, epidemic and modelling terms. We will apply a search algorithm in each database as follows: (“mathematical” OR “dynamical” OR “deterministic” OR “stochastic” OR “Fuzzy”) AND (“co-infection” OR “co-dynamics” OR “dual infection” OR “co-circulation”) AND (“epidemiological” OR “epidemic” OR “infectious disease” OR “transmission”) AND (“model” OR “modeling” OR “modelling”). A summary of the search criteria is provided in table 1, and a detailed version for each database is available in the search strategy (online supplemental file 1).
Supplemental material
The search algorithm to retrieve viral-bacterial co-infection models
Study selection
The studies found via the search algorithm across the considered databases will be assembled in the EndNote (V.20.4.1) referencing tool and further imported to Rayyan (https://www.rayyan.ai/), an online systematic review web tool.32 This web systematic review tool is designed to help reviewers screen titles and abstracts, reduce the risk of bias and aid the data extraction process.33 Three reviewers will initially screen the titles and abstracts in accordance with the eligibility criteria to select potential studies that meet the inclusion threshold. Discrepancies on inclusion such as (1) both reviewers are unsure, (2) one reviewer recommends while the other is unsure and (3) one reviewer recommends inclusion while the other recommends exclusion will be resolved through discussion with the help of a third reviewer to reach consensus. The PRISMA diagrammatic structure of the study selection outline will be applied. The next phase involves a review of the full texts and data extraction for selected studies.
Quality assessment and risk of bias
The assessment of the risk of bias in a dynamical co-infection epidemic model may seem difficult to assess due to a lack of measurement for bias in models. Considerations of modelling approach and type of models are the main recognisable features.34 The quality of the selected co-infection modelling articles will be assessed for good dynamical modelling standard practice, referring to guidelines35–37 highlighting the components of an epidemiological model which include model formulation, description, data fitting, evaluations, simulations, analysis, validation and output corroboration.
Data extraction process
Data extraction will be done using a standard Microsoft Excel template while applying the standard PRISMA data collection guidelines.38 Data extraction will be done by TKY with the help of two reviewers to verify the quality of the data extracted.
Data items
The following features of the selected articles will be extracted as illustrated in table 2.
Description of data items to be extracted on the co-infection models
Data synthesis strategy
We will provide a summary of the included studies in tabular form and critically assess them using the TRAnsparent and Comprehensive Ecological standard ecological modelling guideline to characterise the models.35 Subsequently, a narrative synthesis of the findings will be provided. The summary will highlight the following key co-infection model features: modelling approaches, assumptions, parameter estimates and simulations, intervention strategies, the impact of co-infections, model validation and sensitivity analysis, research gaps and opportunities for future studies. Due to insufficient data for meta-analysis, this systematic review will use the SWiM (Synthesis Without Meta-analysis) guidelines39 in its narrative synthesis.
Ethics and dissemination
Ethics approval is not required for a systematic review since it will be based on published work. The output of this study will be submitted for publication in a peer-reviewed epidemiology journal.
Ethics statements
Patient consent for publication
Acknowledgments
The authors thank Edith Cowan University, School of Science, for providing administrative support.
References
Footnotes
X @EAfrifaYamoah
Contributors TKY was responsible for the conceptualisation and design of the protocol, development of the search strategy, design of the data extraction plan, and drafted the manuscript. EA-Y, SR, JC and UM contributed to the supervision and manuscript review and editing. TKY is the guarantor for this study.
Funding This study was supported by Edith Cowan University Higher Degree by Research Fund through the postgraduate research scholarships.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.


