Article Text
Abstract
Objective Despite extensive exploration of potential biomarkers of cardiovascular diseases (CVDs) derived from retinal images, it remains unclear how retinal images contribute to CVD risk profiling and how the results can inform lifestyle modifications. Therefore, we aimed to determine the performance of cardiovascular risk prediction model from retinal images via explicitly estimating 10 traditional CVD risk factors and compared with the model based on actual risk measurements.
Design A prospective cohort study design.
Setting The UK Biobank (UKBB), a prospective cohort study, following the health conditions including CVD outcomes of adults recruited between 2006 and 2010.
Participants A subset of data from the UKBB which contains 52 297 entries with retinal images and 5-year cumulative incidence of major adverse cardiovascular events (MACE) was used. Our dataset is split into 3:1:1 as training set (n=31 403), validation set (n=10 420) and testing set (n=10 474). We developed a deep learning (DL) model to predict 5-year MACE using a two-stage DL neural network.
Primary and secondary outcome measures We computed accuracy, area under the receiver operating characteristic curve (AUC) and compared variations in the risk prediction models combining CVD risk factors and retinal images.
Results The first-stage DL model demonstrated that the 10 CVD risk factors can be estimated from a given retinal image with an accuracy ranging between 65.2% and 89.8% (overall AUC of 0.738 with 95% CI: 0.710 to 0.766). In MACE prediction, our model outperformed the traditional score-based models, with 8.2% higher AUC than Systematic COronary Risk Evaluation (SCORE), 3.5% for SCORE 2 and 7.1% for the Framingham Risk Score (with p value<0.05 for all three comparisons).
Conclusions Our algorithm estimates the 5-year risk of MACE based on retinal images, while explicitly presenting which risk factors should be checked and intervened. This two-stage approach provides human interpretable information between stages, which helps clinicians gain insights into the screening process copiloting with the DL model.
- health informatics
- biotechnology & bioinformatics
- ophthalmology
Data availability statement
Data are available upon reasonable request. The UK Biobank dataset was obtained from UK Biobank (application number 62399). Data can be accessed from the UK Biobank under research agreement (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). Other researchers can apply for UK Biobank data to answer specific research questions.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
STRENGTHS AND LIMITATIONS OF THIS STUDY
Retinal images have been used to predict cardiovascular disease (CVD) outcomes; we explored their use as estimators of 10 traditional CVD risk factors to enhance model explainability in predicting major adverse cardiovascular events within 5 years.
The training process is interpretable by explicitly estimating 10 CVD risk factors, achieving high prediction accuracy.
The two-stage network setup provides clinicians with deeper insights, allowing for the identification of potential algorithmic abnormalities.
Estimating the 10 individual traditional CVD risk factors enables personalised CVD risk profiles and corresponding intervention to reduce CVD risk.
The main limitation is that deep learning methods require high image quality to achieve high model accuracy.
Introduction
Cardiovascular disease (CVD) is the leading cause of death, and it alone caused 17.9 million deaths in 2019 representing 32% of global mortalities.1 Traditionally, risk assessment for CVD disease has been performed based on multiple factors, namely, older age, male gender, hypertension, dyslipidaemia, diabetes, obesity and smoking habit. This strategy developed clinical CVD risk assessment algorithms, such as Systematic COronary Risk Evaluation (SCORE),2 Framingham Risk Score,3 WHO guidelines4 or Pooled Cohort Equations (PEC),5 to provide a quantitative risk measure. To use such tools essentially requires blood samples for glucose, cholesterol and triglycerides levels.
Novel biomarkers of CVD have been sought extensively to improve the assessment to explain what has been known as the residual risk. One of the unique candidates of CVD risk biomarkers is the retinal image assessment. The retina has been recognised as a window to the body, especially for its uniqueness to visualise vascular structures in vivo. It thus has been recognised as a non-invasive screening modality to evaluate organ damage due to hypertension and other cardiovascular (CVD) risks. It has been demonstrated that qualitative assessment of retinal signs can provide CVD risk assessment,5 then quantitative retinal vessel morphological changes,6–8 and more recently, a deep learning (DL) model using retinal images as a sole input to estimate CVD risk.9–11
However, it is still not clear how the retinal image reflects CVD risk factors, and how the assessment results based on the retinal image may be used or interpreted for life-style modification to realise a clinically useful screening. Therefore, we aimed to develop an explainable machine learning method that analyses retinal images to estimate the 5-year risk of the major adverse cardiovascular events (MACE) via explicitly estimating traditional CVD risk factors. In our proposed novel algorithm, we aimed to predict 10 CVD risk factors using a multitask learning structure to capture the internal correlation among risk factors and make predictions for individual risk factors given only retinal images. Then the 5-year MACE risk is estimated in the second stage. Through the first stage, we can make a risk profiling that tells why an individual is predicted to be in higher CVD risk.
Methods
Dataset
Our dataset is acquired from UK Biobank (UKBB) (https://www.ukbiobank.ac.uk/), which contains data from 502 419 UK participants (as of 23 October 2021). We created a subset from the UKBB dataset with 52 297 patients that comes with the complete set of required attributes and supplementary data, which includes age, sex, smoking habits, systolic blood pressure (SBP), diastolic blood pressure (DBP), hemoglobin A1c (HbA1c), high-density lipoprotein (HDL) cholesterol, low-density lipoprotein (LDL) cholesterol, triglycerides, body mass index (BMI) and presence of diabetes, at least one retinal eye image and MACE outcome. A diagram illustrates the flow of data is shown in online supplemental figure 1. We randomly split the dataset into 3:1:1 as training set (n=31 403), validation set (n=10 420) and testing set (n=10 474). There are 327 (1.04%), 119 (1.14%) and 132 (1.26%) incident MACE in the training, validation and testing set, respectively. The risk score distribution for Pooled Cohort Equation, Framingham Risk Score, SCORE and SCORE 2 is shown in online supplemental figure 2. The code to implement the Cohort Equation, Framingham Risk Score, SCORE and SCORE 2 algorithm is available in online supplemental codes 1–4.
Supplemental material
Definition of MACE
To define MACE outcome, we follow the instruction from McQueenie et al12 to extract 5-year incident MACE based on the definition of stroke and myocardial infarction hospitalisation event data. In brief, the International Classification of Diseases, 10th revision mortality codes: ‘I00-I78’, ‘G45’, ‘G451-G454’, ‘G456’, ‘G458’, ‘G459’ and ‘G460-G468’ are used to identify the MACE occurrence in the dataset within 5-year range. A summary of the characteristics of patients in the dataset is shown in table 1.
Summary of characteristics of patients in the UK Biobank dataset
Proposed MACE prediction models and comparisons to the six state-of-the-art algorithms
XMACE model
We propose ‘XMACE’ model which is a two-stage DL model that takes a retinal image as input and estimates CVD risk factors in the first stage and then predicts MACE outcome based on the estimated CVD risk factors in the second stage (figure 1A).
(A) Diagram of XMACE: two-stage MACE prediction network structure. (B) Diagram of XMACE+: MACE prediction network structure with both image and CVD risk factor as inputs. BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; HbA1c, haemoglobin A1c, HDL, high-density lipoprotein; LDL, low-density lipoprotein; MACE, major adverse cardiovascular events; SBP, systolic blood pressure.
The first stage of the XMACE model is a multitask learning model equipped with an ImageNet pretrained ResNet13 as a backbone to estimate 10 CVD risk factors (age, gender, smoking habits, BMI, SBP, DBP, HbA1c, HDL cholesterol, LDL cholesterol and triglycerides) from a retinal fundus image. The retinal images are centre cropped, resized to 480×480 resolution and then augmented with a series of random transformations. The ResNet backbone outputs a 512×1 vector, which is used as an intermediate layer that connects to 10 independent multilayer perceptron modules to estimate risk factors. This structure has advantage of reducing the risk of overfitting by an order of N, where N is the number of tasks.14 Given that the CVD risk factors are not strictly independent, they are correlated with each other in some degree. In this proposed model, a sophisticated intermediate layer can capture such correlation from the multitask training and using it to improve the overall estimation performance.
The second stage of the XMACE model consists of two fully connected layers for binary 5-year MACE classification based on estimated CVD risk factors from the first stage. To visualise the attention region in the image, we deployed a modified version of IGOS algorithm.15
Our model is trained with 4 Tesla V100 32G GPUs with a batch size of 256. The learning rate is set to 0.001 for the first 10 epochs, then reduced to 0.0001 for the additional 50 epochs. The momentum is set to 0.9. The binary cross-entropy (BCE) loss is used to evaluate the gender, smoke habits and MACE prediction. Mean squared error loss is applied to evaluate the rest of the CVD risk factors. Due to the heavy imbalance of our dataset, a weighting function is applied to the BCE losses. The training process is conducted in two steps. The multitask learning in the first stage was trained first, then we freeze its weight and continue to train the MACE classification model in the second stage.
XMACE+ model
We additionally built extended version of XMACE for comparison, namely ‘XMACE+’, to fully use information from both retinal and 10 actual CVD risk factors. XMACE+ uses a retinal image to estimate 10 risk factors in the first stage, which are then combined with actual CVD risk factors. We acquire the pretrained RestNet backbone and modify it to output 512×1 vector 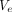 . Similarly, the CVD risk factors from the UKBB dataset are input into a fully connected layer to create a 512×1 vector
. Similarly, the CVD risk factors from the UKBB dataset are input into a fully connected layer to create a 512×1 vector 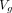 . These two vectors
. These two vectors 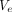 and
and 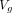 are concatenated to form a new 1024×1 vector
are concatenated to form a new 1024×1 vector 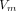 followed by a fully connected layer with 256 neurons and a binary classification layer.
followed by a fully connected layer with 256 neurons and a binary classification layer.
Patient and public involvement
This study was conducted under the UKBB project, with written informed consent obtained from all participants. Patients were not involved in the development of the research question, the design of the study or the outcome measures. Consequently, the research was not directly informed by patients’ priorities, experience or preferences. The allocation of patient data into training, validation and testing sets was done randomly after data collection, without direct patient involvement. As the intervention did not impose any significant burden on patients, there was no assessment conducted by the patients themselves regarding the burden. The deidentified results of this study are publicly released, and there is no specific plan for direct dissemination to the study participants. Finally, since patients were not involved in the recruitment, conduct or advisement of this study, no patient advisers were acknowledged in the contributorship statement or acknowledgments.
Comparison against six state-of-the-art Algorithms in MACE prediction
Our proposed models of XMACE and XMACE+ were compared with the six state-of-the-art algorithms, namely, SCORE, PCE,16 Framingham Risk Score, a logistic regression model, a neural network model and a retinal image-based end-to-end DL model by Poplin et al.10 Detailed model descriptions are presented below. SCORE and the Framingham Risk Score are the two most widely used methods to conduct cardiovascular risk estimation, and we use them as a baseline. The logistic regression and neural network models are adopted in our comparison as they are two of the most widely used statistical methods. The retinal image-based end-to-end DL model was implemented based on the structure proposed by Poplin et al.
SCORE: SCORE is a large-scale study that is based on 12 European cohort studies with 250 000 patients and approximately 3 million person-years of observation. There are 7000 fatal CVD events observed in the dataset. The SCORE conducts CVD risk assessment based on gender, age, LDL, HDL, triglycerides, SBP and smoking status. The model parameters for low-risk region are selected as the UKBB dataset is from the low-risk region. Additionally, we also included the SCORE 217 model in our comparison.
PCE: The PCE are used to estimate a person’s 10-year risk of developing atherosclerotic CVD. PCE conduct CVD risk assessment based on gender, age, LDL, HDL, SBP, diabetic status, race and smoking status. In our experiment, we set all race to Caucasian for UKBB dataset.
Framingham Risk Score: Framingham Risk Score applies Cox proportional hazards regression to evaluate the risk of developing a first CVD event in 8491 Framingham study participants who attended a routine examination between 30 and 74 years of age and were free of CVD. It incorporates age, total and HDL cholesterol, SBP, treatment for hypertension, smoking and diabetes status as risk factors in the calculation. In our case, the UKBB dataset does not have information on treatment for hypertension, so all patients are assumed to be hypertension free.
Logistic regression: the standard logistic regression is applied in this work, where the input are 10 CVD risk factors from the UKBB dataset and the output is the incident MACE binary label.
Neural networks: a neural network with two linear layers is implemented with 512 neurons in the hidden layer. It takes 10 CVD risk factors as input and gives a binary classification result of incident MACE.
Retinal image-based end-to-end DL model: Poplin et al suggested that the inception network can be used to estimate MACE. Following the information provided in Poplin’s work, we implemented a 27-layer inception network.18 The input resolution is 299×299 with a batch size of 256. The learning rate is set to 0.001 for the first 10 epochs, then it is changed to 0.001 for the additional 50 epochs. The momentum is set to 0.9. The BCE loss is used for training, where a weighting function is applied to adjust the imbalance label distribution in the dataset.
Statistical analysis for model evaluation
We conducted three statistical analysis to compare with the state-of-the-art algorithms. We conducted two sets of evaluations. The first evaluation focuses on measuring the accuracy of our XMACE model’s risk factor estimation. The second evaluation is the MACE prediction performance comparisons between our models and the state-of-the-art algorithms, where the area under the receiver operating characteristic curve (AUC) and its 95% CIs are calculated as performance metrics. The 95% CI is calculated based on 2000 bootstrap samples. We calculated Net Reclassification Index (NRI) and Integrate Discrimination Index (IDI) to evaluate the performance between risk prediction models.5 NRI evaluates the improvement in prediction performance gained by adding a marker to a set of baseline predictors. The pairwise value of positive or negative and total NRI shows relationships between different methods. IDI measures the change of the discrimination slope. It is basically the sum of integrated sensitivity and integrated specificity.
Results
Evaluation of risk factor estimation in the first stage in the XMACE model
We followed an approach to Poplin et al10 in terms of error margin setting. The estimation performance is shown in table 2. In terms of age, it achieved 5 years of error margin of 78.3% accuracy; smoke habit and gender achieved 89.8% and 79.6% accuracy, respectively. The distribution of ground truth versus estimated values can be found in figure 1B, where the red diagonal line is drawn to indicate that a prediction and its ground-truth coincide. The red line goes through the area with the highest density, which indicates errors in our model are small. In the additional experiment using the blood vessel mask with red, green and blue (RGB) images to explore any potential gain with additional information, we found that attaching the blood vessel binary mask to the RGB image or replacing the blue channel did not improve the estimation. More visualisation of the model can be found in online supplemental figures 3 and 4.
10 cardiovascular disease risk factors estimated from the retinal image
Evaluation of XMACE prediction performance against the six state-of-the-art algorithms
We compared our algorithm with six state-of-the-art algorithms described in the previous section. Among the models using 10 CVD risk factors based on blood test (table 3), the score-based algorithms, SCORE, SCORE 2, PCE and Framingham Risk Score, achieved the AUCs of 0.682, 0.714, 0.695 and 0.689, respectively. We found that both logistic regression and neural network models obtained higher AUC of 0.758 and 0.763 than the score-based methods. Among the models using predicted 10 CVD risk factors, XMACE+, which additionally uses a retinal image in the second stage, performed the best with AUC of 0.769 (95% CI 0.742 to 0.795).
5-year MACE prediction model performance with or without blood test
Our XMACE model (AUC 0.738, 95% CI 0.710 to 0.766) outperformed the model proposed by Poplin et al.10 (AUC 0.662, 95% CI 0.632 to 0.694) by a margin of 11.5% (95% CIs did not overlap). The NRI value shows that SCORE, Framingham Risk Score and Poplin et al model are overestimating 5-year MACE risk compared with XMACE. The logistic regression and neural network models’ total NRI and IDI values show higher similarity to our XMACE model (shown in table 4).
Model comparison
Discussion
Overall, XMACE+, the model using a retinal image and measured 10 CVD risk factors, obtained the highest AUC among all, which indicates the maximum performance can be achieved when given both actual risk factor and retinal image to the DL model. At the same time, we found that XMACE, the mode using a retinal image as a sole input, performed the best within the models without using blood test results. Our results indicate the potential of using a DL model with retinal fundus images as a tool for rapid, non-invasive MACE risk analysis. Our XMACE model is purely based on retinal images; it first estimates CVD risk factors to predict MACE. Our model showed superior performance (0.738 AUC) over the traditional score-based methods2 3 17 (0.682, 0.714 and 0.689 AUC, respectively) as well as the state-of-the-art DL model proposed by Poplin et al.10 (0.662 AUC). In terms of cardiovascular CVD risk factors estimation from retinal images, our model obtained 78.3% of age predictions fells into the 5 years error margin; smoke habit and gender prediction accuracy were 89.8% and 79.6%, respectively. This is also consistent with previous studies that suggest retinal imaging6 8 19–21 contains information about cardiovascular CVD risk factors and MACE.
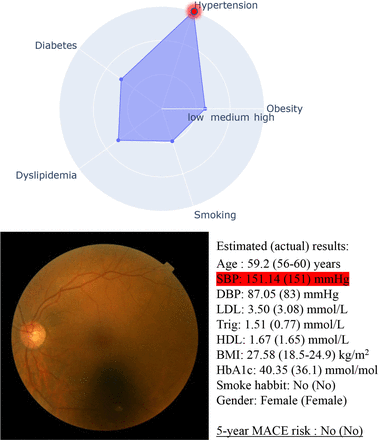
Our experiments also showed XMACE+ performed the best; however, XMACE+ takes a retinal image and 10 actual risk factors as input. Meanwhile, XMACE requires merely retinal imaging without any additional personal information and blood tests. We consider this can fit into a fast screening before doing blood tests. It has potential applications in senior homes, community centres and local pharmacies as a non-invasive quick health monitoring tool. Another advantage of XMACE is its excellent interpretability. Our two-stage approach outputs CVD risk factors in the first stage, which increased the explainability to allow researchers to have some level of confidence in the second stage MACE prediction. Figure 2 is an example of the retinal images and estimated CVD risk factors. Even though each subject has similar 5-year MACE risk, they have different risk profiles that require matched personalised intervention for preventing CVD. By explicitly estimating the interpretable CVD risk factors, XMACE has the potential to generate lifestyle recommendations to patients, which is shown in figure 3 (more examples can be found from online supplemental figures 5–8). The classification of risk level is available in online supplemental table 1.
Ground truth versus estimation on 10 risk factors; the colour indicates the density of samples. BMI, body mass index; DBP, diastolic blood pressure; HbA1c, haemoglobin A1c, HDL, high-density lipoprotein; LDL, low-density lipoprotein; SBP, systolic blood pressure.
Cardiovascular disease (CVD) risk profiling based on the retinal image: a case with high risk of CVD due to hypertension. BMI, body mass index; DBP, diastolic blood pressure; HbA1c, haemoglobin A1c, HDL, high-density lipoprotein; LDL, low-density lipoprotein; MACE, major adverse cardiovascular events; SBP, systolic blood pressure.
Despite the promising results, our study has several limitations. First, the UKBB dataset contains different levels of image qualities. Similar to Poplin et al, we deployed a simple algorithm to filter out low-quality images. The source of low-quality images may be from improper use of equipment or seniors with chronic conditions deteriorated their retinal images. Future work could be creating an algorithm that identifies such low-quality images caused by chronic conditions. Additionally, tailoring an image enhancement algorithm for low-quality images could be an interesting direction. The second limitation is that some important CVD risk factors, such as HbA1c, BMI and triglycerides, are missing from many patients in the original UKBB dataset, which reduces the number of available training data (out of 502k patients in the UKBB dataset, only 52.3k patients satisfy the criterial in this study). A semisupervised learning algorithm can be applied in this situation that conducts training on both labelled and unlabelled data. Using both labelled and unlabelled, data will largely increase the pool of data available for training, which potentially increases training quality. There have been studies22 demonstrating potential newer risk factors other than the traditional CVD risk factors we used in this study. However, there has been no consensus if they are good enough to improve the performance in the clinical setting. Our experiments also found lipids estimate accuracy is worse than age and blood pressure which implies the lipids has higher volatility on person’s health conditions which is has less direct effect on the retinal.
In summary, we have provided evidence that a DL model with retinal image input may reveal hidden information in the image that allows better MACE prediction than traditional risk models while securing explainability by providing estimated CVD risk profile. Our proposed model coupled with a mobile retinal imaging device has the potential to realise fast and simple CVD screening that does not require blood test facilities. Further clinical validation with a prospective cohort will secure our findings in this context.
Data availability statement
Data are available upon reasonable request. The UK Biobank dataset was obtained from UK Biobank (application number 62399). Data can be accessed from the UK Biobank under research agreement (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access). Other researchers can apply for UK Biobank data to answer specific research questions.
Ethics statements
Patient consent for publication
Ethics approval
This study involves human participants. The UK Biobank obtained ethics approval from North West–Haydock Research Ethics Committee (reference: 21/NW/0157/299116). Our study received ethics approval from the Research Ethics Committee of the Osaka University Hospital (reference: 22234(T1)). Our study followed the principles of the Declaration of Helsinki. Participants gave informed consent to participate in the study before taking part.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Contributors YQ and RK contributed to the design and analysis of the study and drafted and revised the manuscript according to other authors’ comments. LL, YN, HN and KN contributed to the design of the study, made critical comments on the manuscript and revised the paper critically. All authors approved the final version of the manuscript. RK is responsible for the overall content as guarantor.
Funding This work was supported by Council for Science, Technology and Innovation, Cross-ministerial Strategic Innovation Promotion Program, ‘Innovative AI Hospital System’ (Funding Agency: National Institute of Biomedical Innovation, Health and Nutrition, SIPAIH20D03). This work was also supported by JSPS KAKENHI, grant number 19K10662.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.