Article Text
Abstract
Objectives To evaluate the risk of exposure to SARS-CoV-2 in naturally ventilated hospital settings by measuring parameters of ventilation and comparing these findings with results of bioaerosol sampling.
Study design Cross-sectional study.
Study setting and study sample The study sample included nine hospitals in Dhaka, Bangladesh. Ventilation characteristics and air samples were collected from 86 healthcare spaces during October 2020 to February 2021.
Primary outcome Risk of cumulative SARS-CoV-2 infection by type of healthcare area.
Secondary outcomes Ventilation rates by healthcare space; risk of airborne detection of SARS-CoV-2 across healthcare spaces; impact of room characteristics on absolute ventilation; SARS-CoV-2 detection by naturally ventilated versus mechanically ventilated spaces.
Results The majority (78.7%) of naturally ventilated patient care rooms had ventilation rates that fell short of the recommended ventilation rate of 60 L/s/p. Using a modified Wells-Riley equation and local COVID-19 case numbers, we found that over a 40-hour exposure period, outpatient departments posed the highest median risk for infection (7.7%). SARS-CoV-2 RNA was most frequently detected in air samples from non-COVID wards (50.0%) followed by outpatient departments (42.9%). Naturally ventilated spaces (22.6%) had higher rates of SARS-CoV-2 detection compared with mechanically ventilated spaces (8.3%), though the difference was not statistically significant (p=0.128). In multivariable linear regression with calculated elasticity, open door area and cross-ventilation were found to have a significant impact on ventilation.
Conclusion Our findings provide evidence that naturally ventilated healthcare settings may pose a high risk for exposure to SARS-CoV-2, particularly among non-COVID-designated spaces, but improving parameters of ventilation can mitigate this risk.
- Airborne transmission
- COVID-19
- SARS-CoV-2
- ventilation
- healthcare
- hospital
- nosocomial infection
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. All data are included within the manuscript and supplementary files.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
A major strength of this study that it provides an assessment of risk for airborne SARS-CoV-2 transmission in naturally ventilated healthcare facilities based on parameters of ventilation as well as direct detection of viral RNA across a range of healthcare settings.
Another strength of this study is that it used low-cost carbon dioxide metres to assess ventilation, which could be replicated in other resource-limited settings to measure ventilation and inform risk mitigation efforts of airborne diseases.
Additionally, we used an elasticity analysis to identify the parameters of ventilation that have the greatest impact on improving ventilation per interval change.
One limitation is the cross-sectional nature of this study, allowing us to only measure SARS-CoV-2 RNA at a single time point, which may not fully represent longitudinal risk.
Another limitation is that we did not confirm virus viability because we did not have access to viral culture.
Background
Hospital-associated exposure to SARS-CoV-2 was a critical driver of disease spread early in the pandemic.1 Mitigation efforts in healthcare facilities initially focused on reducing droplet and contact exposure to SARS-CoV-2.2 However, airborne transmission has been increasingly recognised as an important route of virus spread.3–6 A key component towards reducing the spread of aerosol-transmitted diseases is to ensure adequate ventilation.7 Many hospitals in high-resource settings isolate patients with COVID-19 in negative pressure rooms with regulated rates of air exchange. These advanced engineering systems are often not available in resource-constrained settings where patients are commonly kept in communal wards that are reliant on natural ventilation. Very little is known about the risk for airborne SARS-CoV-2 transmission in these settings.
The large size of droplet particles limits their spread in both space and time, requiring close proximity to an infected individual to establish exposure. Aerosols, in contrast, can travel in suspended air plumes with prolonged viral persistence.8 9 Exposure to SARS-CoV-2 through aerosols can, thus, occur over larger space and time parameters, posing greater cumulative risk in shared spaces with air recirculation and/or inadequate ventilation. Several studies have detected SARS-CoV-2 RNA in air samples from hospitalised patients with COVID-19, including up to 13 feet away from infected patients, demonstrating that the virus is carried in aerosols.10–17 Two studies used culture to demonstrate the presence of viable virus in aerosol samples, further supporting airborne transmission as a potential pathway of exposure.10 18
With the ongoing transmission of SARS-CoV-2 globally, governments have struggled to protect one of the most vulnerable populations: healthcare workers. Large numbers of healthcare worker infections have further burdened healthcare systems, particularly in low and middle-income countries (LMICs), where dire shortages of healthcare workers preceded the COVID-19 pandemic.19–21 Adequately protecting healthcare workers relies on understanding transmission risk to inform risk mitigation strategies—from designing engineering controls to implementing appropriate personal protective equipment (PPE) recommendations. Aerosolized particles produced in such environments without adequate ventilation mechanisms could be a critical exposure pathway for healthcare providers. A more complete understanding of the risk of airborne SARS-CoV-2 transmission is imperative for developing policies and practices that improve the safety of healthcare facilities.
WHO guidelines for natural ventilation for infection control in healthcare settings recommend a ventilation rate of 60 L/s per person (L/s/p) for general wards and outpatient departments (OPD) and a ventilation rate of 160 L/s/p in the setting of aerosol-generating procedures (AGP) to prevent airborne infections.22 Obtaining these ventilation parameters depends on ensuring adequate air exchange efficiency, which is dictated by factors such as opening area to outside, cross-ventilation and person density.23 24 A useful metric for approximating ventilation in a steady-state, naturally ventilated indoor environment is by measuring CO2, which reflects the amount of air that is exhaled breath based on the number of people in a given space.25 This can be translated into infectious risk for airborne infections when the pathogen abundance in the environment can be estimated or modelled.26 27
The objective of this study was to quantify the potential risk of SARS-CoV-2 infection across naturally ventilated healthcare spaces in Dhaka, Bangladesh and to compare these findings with SARS-CoV-2 RNA detection in aerosols in those spaces. We compared this against SARS-CoV-2 RNA detection in mechanically ventilated healthcare spaces. We also evaluated the association between ventilation and the presence of SARS-CoV-2 RNA and analysed drivers of ventilation to identify potential factors to modify the risk in naturally ventilated spaces.
Methods
Patient and public involvement
No patients were directly involved in the study.
Study setting
We collected ventilation measurements and conducted environmental bioaerosol sampling in six public and three private hospitals in Dhaka, Bangladesh between October 2020 and February 2021. We selected naturally ventilated rooms for sampling, which were categorised by whether patients in that area were known or suspected to have COVID-19 or not. We also sampled mechanically ventilated spaces for comparison. We included a range of room types across facilities, including open wards, cabins (semiprivate rooms), intensive care units (ICUs), OPDs, PPE doffing areas and bathrooms.
Data collection
We collected measures of ventilation in each of the naturally ventilated environments to use in our risk modelling. We used a handheld carbon dioxide metre (Extech, Boston, Massachusetts) to assess levels of CO2 in parts per million (ppm) at 5 min intervals across the 30 min sampling period as well as temperature and humidity. We averaged the CO2 levels across the sampling period to approximate a steady-state concentration. We also collected outdoor CO2 measurements at the beginning of each day of sample collection. We used these values to calculate the absolute ventilation (L/s) per sampling space using the following equation:25

where G is equal to the average CO2 generation rate per person, n is equal to the number of people in the space,  is equal to the averaged CO2 measurements during the sampling period and
is equal to the averaged CO2 measurements during the sampling period and  is equal to the outdoor CO2 measurement taken in the morning of each sampling day. Ventilation rate (L/s/p) was calculated by dividing the absolute ventilation by the number of people in the space. We also calculated air changes per hour (ACH) using the equation:
is equal to the outdoor CO2 measurement taken in the morning of each sampling day. Ventilation rate (L/s/p) was calculated by dividing the absolute ventilation by the number of people in the space. We also calculated air changes per hour (ACH) using the equation:
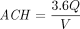
where V is equal to the room volume (m3).
We also collected information on the number of patients with COVID-19 (confirmed or suspected) and without COVID-19, healthcare staff and visitors present throughout the sample collection time to inform our risk models. For our ventilation analysis, we used a GLM V.15 Compact Laser Measure (Bosch, Farmington Hills, Michigan) to measure room height, width, length and the area of all open windows and doors. We considered a room to have cross-ventilation if there was a window or door open on two opposing walls. Within each of the spaces, we recorded the proportion of patients wearing face masks.
Risk modelling
We used the modified Wells-Riley equation proposed by Rudnick and Milton to generate the probability of infection in naturally ventilated spaces over a 40-hour time interval to represent risk during an average work week.27

where I is the number of infected individuals in the space, p is the average respiration rate of an adult (6 L/min at rest), t is the time elapsed in an interval and q is the quanta emission rate (QER), which accounts for the number of infectious doses emitted by an individual over a given time.
For I, we counted all patients in designated COVID-19 spaces as potential infectors since a positive COVID-19 test is required for admission to a COVID-designated area. Non-COVID spaces included open wards where all patients have tested negative, and OPDs, where the number of infected individuals is unknown. We set I equal to 1 for non-COVID open wards to represent a hypothetical scenario if one patient was to be incorrectly diagnosed as negative. For OPDs, we assumed 3% of non-staff were infected based on concurrent SARS-CoV-2 test positivity data for the given sampling period (rounding up for fractional values).28
To obtain q, we used activity-specific distributions characterised by Buonanno et al.29 The authors hypothesised that there is uncertainty around the QER because of random variation in the concentration of viral load expired during activity. For ICUs, open wards and cabins, we assumed the activity level was ‘resting, oral breathing’ for potential infectors (log10(QER per hour)~N(−0.429, 0.720)) and for OPDs, we assumed ‘light activity, talking’ for potential infectors (log10(QER per hour)~N(0.698, 0.720)). We applied a Monte Carlo method to draw values for each potential infector QER within each space (qi). Based on studies reporting time to diagnosis and hospitalisation for SARS-CoV-2, we estimated that inpatients were on average 8 days into their disease course compared with 3 days for outpatients.30–32 Since viral shedding decreases with duration of illness, we, thus, subtracted 0.5 (log10 scale) from each qi drawn for inpatients based on estimated differences in viral shedding between day 3 and day 8 of illness.33 34 We calculated the average qi drawn for all potential infectors, took the antilog and multiplied by one minus the proportion of mask-wearing patients (Pm) times the efficacy of surgical masks (E) in preventing outward transmission of infectious aerosols (50%) to obtain the final value of q for each sampling space.35–37

Using the final calculated value of q for each space, we calculated the risk for each sampling space using the aforementioned modified Wells-Riley equation and took the median risk for each type of space (eg, to obtain the median risk for OPDs). We repeated random draws of qi for potential infectors 1000 times to obtain a distribution of 1000 type-specific medians. We calculated the overall median risk by type of space for the 1000 simulations as well as the 2.5th percentile and 97.5th percentile of the generated distribution. We also used the 1000 calculated q values to obtain risk curves for individual sampling spaces.
Bioaerosol sampling
To compare the results of our risk modelling to empirical data, we collected air samples over a 30 min time period in each space and measured SARS-CoV-2 viral copies. We collected air samples from both naturally ventilated and mechanically ventilated spaces to compare risk of SARS-CoV-2 RNA detection.
Sample collection
We used a liquid impinger biosampler with a BioLite Air sampling pump (SKC Ltd., Eighty Four, Pennsylvania) set to a calibrated flow rate of 12.5 L/min. The biosampler was set as close to the centre of each room as possible and 1–1.5 m above the ground. The vessel connected to the liquid impinger was filled with 10 mL of 1× phosphate buffered saline (PBS) (pH 7.4) for bioaerosol collection. Immediately after the collection, we added 7 mL of NucliSENS RNA stabilising lysis buffer (bioMérieux, Durham, North Carolina) and transported the samples at 2°C–8°C. Between each sample collection, we decontaminated the biosampler by separating all components of the device and submerging them into 10% sodium hypochlorite for 15 min followed by rinsing the sampler components three times with distilled water. To rule out any carryover contamination from the previous run, we rinsed the biosampler after every decontamination with 1 mL distilled water that we analysed by RT-qPCR. Additionally, to ensure that there was no backflow contamination by the pump, we collected negative controls daily by attaching an N95 filter to the biosampler inlet over a 30 min sampling period and tested the PBS collection fluid by RT-qPCR.
Sample processing and RNA extraction
Before RNA extraction, we concentrated 14 mL of sample collected in PBS and lysis buffer to 500 µL using Amicon Ultra-15 Centrifugal Filter Units (Milipore Sigma, cat# C7715) at 5000 rpm for 20 min. We extracted RNA using a modified (additional 25 µL of proteinase K was added to the reaction during the lysis step) MagMAX Viral/Pathogen Ultra Nucleic Acid Isolation Kit (Applied Biosystems A42356) as per the manufacturer’s instructions. The RNA was eluted in a 50 µL elution buffer and stored at −20°C until further testing.
Sample analysis
We performed RT-qPCR using the U.S. Centers for Disease Control and Prevention qualified primers (500 nM) and TaqMan probes (300 nM) amplifying N1 and N2 regions of SARS-CoV-2 N-gene.38 TaqPath one-step RT-qPCR mastermix (Invitrogen, Darmstadt, Germany) was used in a 20 µL reaction volume and analysed on a StepOne-Plus (Applied Biosystems) instrument, using the following programme: 10 min at 50°C for reverse transcription, followed by 3 min at 95°C and 40 cycles of 10 s at 95°C, 15 s at 56°C and 5 s at 72°C. We estimated the number of copies per sample from a standard curve using 10-fold serially diluted SARS-CoV-2 synthetic RNA (ATCC Cat#VR-3276SD). All the samples were run in duplicates and averaged. A positive sample was defined as having a Ct value less than 38 in at least two measurements (N1 target positive in both replicates, N2 target positive in both replicates, or positive N1 target + positive N2 target).
Ventilation analysis
To determine which parameters of ventilation had the greatest impact on ventilation in naturally ventilated spaces, we analysed the association between each ventilation parameter measured and log10 absolute ventilation using univariate linear regression models to obtain unadjusted mean differences, excluding extreme outliers from each parameter distribution. To alleviate sample size constraints, we used a least absolute shrinkage and selection operator (LASSO) regression to select parameters for a multivariable model. We used fivefold cross-validation to select the λ penalty at the minimum mean-squared error. Any variables with non-zero coefficient values were included in a multivariable linear regression model to obtain adjusted mean differences and 95% CIs.
We assessed variable importance using elasticity, which standardises the estimated parameters by multiplying the coefficient from the regression model with the mean of the associated variable divided by the mean of the outcome. This results in an estimate of the per cent change in the outcome for a per cent change in the exposure and makes the parameters comparable in a postestimation step, while keeping the regression coefficients in units that are understandable.39

where dY/dX is the slope of Y with respect to X for a given X, multiplied by the ratio of a given value of X to a given value of Y (we used the mean of both). We performed this for the point estimates and associated CIs from our multivariable regression to identify variables that most influenced log10 absolute ventilation. To assess risk stratified by important variables, we employed the approach detailed above and performed 1000 simulations on each space to generate sample-specific risk medians.
Results
We sampled a total of 86 locations, including 28 open wards, 9 ICUs, 18 cabins, 12 OPDs and 19 other spaces, including bathrooms, PPE doffing rooms, COVID-19 testing areas and a canteen. Sixty-two (72%) of the spaces were naturally ventilated and 24 (28%) were mechanically ventilated. Among the 86 spaces, 65 (76%) were areas with patients confirmed or suspected to have COVID-19, and 21 (24%) were non-COVID areas. The areas with patients with COVID-19 had fewer people (median: 6) compared with non-COVID areas (median: 38) (p value <0.001). Within OPDs, the median per cent of people found to be wearing masks was 75% (range: 31%–100%) with the vast majority wearing surgical masks. No hospitalised patients were observed to be wearing masks. Healthcare staff in COVID-19 ICUs wore N95s while staff in other settings typically wore either surgical masks or no masks.
Of the naturally ventilated spaces, OPDs had the highest person density of the spaces sampled (0.45 people/m2), followed by open wards (0.19 people/m2) (table 1). Most rooms apart from wards and cabins did not have open windows. However, the majority of sampled spaces had at least one open door at the time of sampling. Forty per cent of the spaces had evidence of cross-ventilation. Fans were used for climate control in nearly half of the rooms, while ICUs predominantly relied on wall-mounted or portable air conditioning units.
Descriptive parameters of naturally ventilated spaces (n=62)
Among the naturally ventilated spaces, the median CO2 level was 729 ppm (range: 410–1936 ppm) (table 1). CO2 values were fairly steady over the 30 min sampling period (online supplemental figure 1). The median absolute ventilation among spaces where ventilation was able to be calculated (excluding five spaces with no people present during the sampling period) was 196 L/s (IQR 114–530) and the ventilation rate was 15.6 L/s/p (IQR 10.9–42.5) (table 1). An overwhelming majority (78.7%) of rooms had ventilation rates that fell short of the recommended ventilation rate of 60 L/s/p (figure 1).
Ventilation rates across naturally ventilated spaces (n=57), excluding five spaces where no people were present at the time of sampling. Dashed line indicates the recommended ventilation rate of 60 L/s/person. Green dots (n=10) signify sampled locations above the 60 L/s/person threshold; yellow dots represent ventilation rates of 50–60 L/s/person (n=3); light orange dots represent 30–50 L/s/person (n=6); dark orange dots represent 10–30 L/s/person (n=27); and red dots are below 10 L/s/person (n=11). ICU, intensive care unit.
Estimating infectious risk
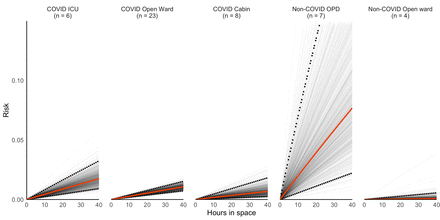
On average, COVID-19 ICUs and COVID-19 open wards had the highest number of potential infectors (median=6 for both), compared with COVID-19 cabins (median=2) and OPDs (median=1). However, we found that among the five types of patient care spaces sampled, OPDs were overall the highest risk location (figure 2). After 40 hours in OPDs, the median risk of infection in the absence of other mitigation measures was 7.7% (95% CI 2.2% to 25.2%). ICUs designated as COVID-19 spaces were the second riskiest spaces, with 1.8% risk (95% CI 0.91% to 3.2%) over 40 hours. Cabins and open wards for patients with COVID-19 had a similar risk profile (0.69%; 95% CI 0.25% to 1.82% compared with 1.1%; 95% CI 0.8% to 1.5%). Open wards that were not designated for patients with COVID-19 carried the least risk under the assumed scenario of one potential infector (0.08%; 0.01% to 0.56%).
Risk of SARS-CoV-2 infection over a 40-hour time period, by type of sampling space. Grey lines are simulation-specific averages of the median risk by type of sampling space. The solid red line is the overall median risk of infection over time and the dashed black lines are the 2.5th and 97.5th percentile. ICU, intensive care unit; OPD, outpatient department.
SARS-CoV-2 RNA detection
Of the 86 bioaerosol samples tested by RT-qPCR, we detected SARS-CoV-2 RNA in 16 (18.6%) of the samples (table 2). SARS-CoV-2 was detected in 22.6% of naturally ventilated spaces and 8.3% of mechanically ventilated spaces, though this difference was not statistically significant (p=0.128). The room types with the highest proportion of positive samples were non-COVID wards (2/4; 50.0%) and OPDs (3/7; 42.9%). Among positive samples, the median copy number was 189 (range: 79–929). There was no difference in the median copy number between COVID-19 and non-COVID spaces, where SARS-CoV-2 was detected (p=0.336).
SARS-CoV-2 RNA RT-qPCR positivity and copy numbers in bioaerosol samples collected from nine hospitals in Dhaka, Bangladesh
Ventilation analysis
When analysing the architectural and ventilation features of the naturally ventilated sampling spaces, we found that open door area was the most important parameter associated with absolute ventilation in a multivariable analysis with a 0.74% change in log10 ventilation per 10% change in area (95% CI 0.03% to 1.4%) (table 3, online supplemental figure 2). Total open area of doors and windows combined and average number of people were the next most influential factors; however, these estimates were only marginally significant (0.52%; 95% CI −0.11% to 1.1% and 0.42%; 95% CI −0.04% to 0.88%). Cross-ventilation was less important than these factors but was the only other significant parameter in the multivariable analysis besides open door area (0.4%; 95% CI 0.29% to 0.77%). We found that the risk of SARS-CoV-2 infection was moderated by open areas of doors and whether there were windows on opposite walls (online supplemental figure 3).
Univariable, multivariable and elasticity analysis of ventilation parameters
Discussion
In this study, we demonstrated the potential for airborne transmission of SARS-CoV-2 by revealing inadequate ventilation, a known driver of infectious risk, in a variety of naturally ventilated healthcare spaces. Our main threshold for adequate ventilation was based on the WHO-recommended 60 L/s/p, though AGPs were also likely performed in many of the sampled spaces, including SARS-CoV-2 sample collection and positive pressure ventilation. Only two spaces met the ventilation threshold for AGPs, 160 L/s/person. Overall, there was severely inadequate ventilation across most spaces, indicating a need for large-scale improvements to reduce environmental exposure risk.22
We found that patient entry points to the hospitals (OPDs), where patient COVID-19 status was unknown, had the highest risk for airborne SARS-CoV-2 exposure based on infectious risk modelling, despite high rates of mask wearing. This was corroborated by high rates of viral RNA detection in these spaces. We believe that this is explained by several factors. Patients presenting to OPDs likely have higher viral shedding because they are earlier in their disease course, and they may have a higher level of activity than patients admitted to the hospital, both of which we accounted for in our modelling. This corresponds with findings that the majority of SARS-CoV-2 infections are likely transmitted by people early in their disease course.40 Additionally, although OPDs had higher absolute ventilation than some other areas, it was still insufficient to mitigate the risk of higher viral shedding and higher person density. Conversely, despite high numbers of infectious individuals in COVID-19 wards, high ceilings combined with cross-ventilation likely reduced the overall risk. Diverging from our risk models, there was also a high rate of detection in non-COVID wards, potentially due to imperfect testing methods during triage. The high proportion of positive samples in non-COVID spaces is an important finding because healthcare workers may underestimate exposure risk in these settings.
Areas with patients confirmed or suspected to have COVID-19 are often considered to be high-risk areas for transmission potential, leading to differential PPE recommendations across types of patient care areas.41 Additionally, a lower perceived risk of exposure to SARS-CoV-2 has been associated with decreased adherence to PPE use.42 43 In accordance with this observation, PPE use among healthcare staff in our study appeared to be less stringent in non-COVID areas. Our study revealed a strong dichotomy between perceived risk and actual risk of exposure to SARS-CoV-2 across healthcare spaces, which appears to be heavily modulated by viral emission rate and timing of disease course. These findings demonstrate an urgent need for enhanced ventilation measures across all healthcare settings as well as enhanced PPE recommendations for the protection of healthcare workers, patients and visitors against nosocomial transmission of airborne diseases, including COVID-19.
We demonstrated direct detection of SARS-CoV-2 RNA in a high percentage of healthcare spaces—we found SARS-CoV-2 RNA in 18.6% of samples. In contrast, four other studies failed to detect SARS-CoV-2 RNA in mechanically ventilated healthcare spaces despite obtaining substantially larger volumes of filtered air and placing air samplers more proximal to infected patients.44–47 This may reflect lower risk associated with mechanically ventilated spaces that are better able to filter infectious particles. Similarly, in our study, we found a lower rate of SARS-CoV-2 detection among mechanically ventilated spaces (8.3% vs 22.6%), though this difference was not statistically significant, likely as a result of sample size limitations. We also did not detect SARS-CoV-2 in bathrooms or PPE doffing rooms, contrasting with other studies that found these to be high-risk areas, indicating that risk in these spaces may be context dependent.48
Of the samples in which we detected SARS-CoV-2 RNA, the median copy number was less than 1 copy/L of air sampled, which is comparable to other studies where SARS-CoV-2 RNA was detected in air samples from healthcare settings.10–14 49 However, given that previous estimates have suggested that an infectious dose of SARS-CoV-2 may be in the range of 100–300 virions,50 51 this could indicate that the risk from aerosol transmission may be substantial, especially since the cumulative exposure may be more important for establishing infection than exposure at a single time point.52 Additionally, healthcare providers may interact more closely with patients at times, and ventilation may decrease if windows are closed, such as at night or during inclement weather, potentially increasing exposure risk. Moreover, a previous study was able to culture virus from air samples when the Ct value was less than 38, which was the threshold used for determining positivity of air samples in the present study.10
A unique aspect of this study is an investigation of which parameters of ventilation have the greatest impact on air exchange in naturally ventilated settings and subsequently how risk was modified by these factors. Open areas and cross-ventilation were found to be the most influential parameters in the elasticity analysis, which have been previously demonstrated to be highly effective in improving ventilation.53 While these parameters can often be easily modified, such as by ensuring windows and doors are opened to the fullest extent, enhancements such as skylights can further direct air flow and maximise ventilation.53
Our modelled estimates of risk are likely conservative because we did not take into account parameters around AGPs, which can further increase risk for airborne transmission. In contrast with other studies,12 17 54 the ICUs in our study did not have a high rate of SARS-CoV-2 detection. However, inadequate ventilation in these spaces put them as the second highest risk space in our risk models. Additionally, we assumed 50% effectiveness of masks at preventing outward disease spread by patients based on laboratory studies, though mask use practices (eg, correct mask wearing over nose and mouth) and mask filtration efficiency are likely to be lower in real-world scenarios compared with standardised study conditions. Furthermore, we identified potential infectors in OPD spaces based on an average test positivity rate at the lowest point in the pandemic in Bangladesh,28 which coincided with our air sample collection period. However, the risk of infection with SARS-CoV-2 is likely to increase in these spaces as community transmission increases because a higher proportion of patients seeking care are likely to have COVID-19. Therefore, our estimates are conservative for these settings and actual risk during a given time period may be substantially higher.
One limitation of this study is that viral culture was not available. While assessing for the presence of SARS-CoV-2 using RNA does not address pathogen viability; epidemiologic and laboratory studies have demonstrated the possibility of aerosol transmission in the setting of disease spread without direct contact between individuals.55–58 Additionally, laboratory studies have revealed that the SARS-CoV-2 can remain viable in the air for up to 16 hours with no detectable half-life.9 Furthermore, viability has never been demonstrated in well-known airborne-transmitted diseases such as measles given limitations in collection methods.59
Another limitation is the cross-sectional nature of this study, allowing us to only measure SARS-CoV-2 RNA at a single time point, which can be affected by disease constellations, density and activity of patients. To address this limitation, we combined these direct measures of viral presence with indirect drivers of risk, such as ventilation, to obtain a more complete assessment as ventilation parameters are likely to remain stable over time. This approach to using ventilation parameters as assessed by CO2 levels has been validated as a proxy for risk of transmission of other airborne diseases, including tuberculosis and measles.23 26 27 60
Additional limitations include that CO2 is gas that may distribute throughout a space differently from infectious aerosols. As such, the Wells-Riley model assumes perfect mixing, which is unlikely true of the measured spaces. We also did not directly measure air flow in or out of windows and instead relied on the rebreathed fraction of air to approximate air exchanges. Furthermore, because we relied on CO2 to approximate ventilation, we were unable to assess ventilation among mechanically ventilated spaces. As with all air sample collection devices, the device used in this study may have imperfect efficiency at capturing infectious particles. The manufacturer reports an efficiency close to 100% for particles 1.0 µm in diameter when sampled at 12.5 L/min, though the efficiency decreases to 90% for particles 0.5 µm in diameter.61 Given challenges related to data collection in the field, we may have had suboptimal recovery of viral RNA. Furthermore, the QER proposed by Buonanno et al is based on a number of assumptions about viral loads and emission rates of SARS-CoV-2 in the setting of limited data.29
Given increasing evidence for aerosol transmission of SARS-CoV-2, engineering modifications of healthcare spaces are of utmost importance for reducing risk to healthcare workers, visitors and other patients of healthcare facilities.62 Modifications of healthcare spaces were proposed during the first SARS outbreak, including window exhaust fans, that require minimal infrastructure investment and may be relevant in contexts such as Bangladesh and other LMICs.22 63 Enhanced effects of cross-ventilation may be observed if the open area of windows or doors on opposing walls can be maximised. Another strategy that has been particularly useful at reducing infectious risk of airborne diseases by improving ventilation as well as inactivating pathogens is upper room ultraviolet germicidal irradiation, though additional maintenance may be required to sustain these systems.64 65 Additionally, triaging patients in tents outside hospital facilities or setting up separate fever clinics could result in less virus emission within hospitals. Barring modifications to the environmental context, reducing the number of people in a space and increased enforcement of surgical mask wearing for patients, especially in OPDs, and N95 use among staff in all patient care areas in the setting of community transmission of SARS-CoV-2 may be downstream solutions.62 66
Conclusion
COVID-19 remains an ongoing threat to populations around the world. As with outbreaks of other emerging infectious diseases,67–69 healthcare facilities were a source of transmission in the early spread of SARS-CoV-2, resulting in an excess of healthcare worker infections.1 20 Improving the safety and resiliency of healthcare facilities is imperative for protecting against future epidemic spread. This is only possible by adequately equipping healthcare spaces with durable mitigation measures that are effective against a range of transmission patterns. While the COVID-19 pandemic will eventually subside, the risk of airborne transmission of other diseases remains a substantial risk in healthcare facilities with inadequate ventilation. Now is the critical moment of action to prevent healthcare facilities from further amplifying the current and future pandemics.
Supplemental material
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information. All data are included within the manuscript and supplementary files.
Ethics statements
Patient consent for publication
Ethics approval
The current study was approved by the research and ethics review committees at icddr,b (number PR-20063). Because the study did not involve a human subjects component, it was considered not to require IRB approval at Stanford University or University of California Berkeley.
Acknowledgments
We would like to thank Aninda Rahman, Communicable Disease Control Program, Directorate General of Health Services (DGHS), for his facilitation to conduct this research. We would also like to thank Pedro M. de Oliveira, Savvas Gkantonas and Epaminondas Mastorakos from the University of Cambridge, Department of Engineering for sharing thoughtful insights around airborne transmission risk modeling. Additionally, we appreciate the cooperation of the hospitals in Dhaka who allowed us to collect these measurements.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
AS, CH, KIH, RV, CL, M.OFB, AN, M.GDH, MBA and JRA contributed equally.
Contributors AS contributed to the research conception and design and manuscript writing; CH performed data analysis and contributed to research design and manuscript writing; RV participated in the research design, data analysis and interpretation, and manuscript writing; KIH contributed to the research design, data collection, and data analysis; CL participated in the research design, data analysis, and manuscript writing; AN contributed to research design and data collection; MOFB participated in data collection; MGDH contributed to research design and dissemination of findings for public health practice; MBA contributed to the research conception and design and interpretation of results; JRA contributed to research conception and design, interpretation of findings, and manuscript writing. As the guarantor, AS accepts full responsibility for the work and the conduct of the study, had access to the data, and controlled the decision to publish.
Funding This study was supported by an anonymous donation to the Stanford University School of Medicine. icddr,b acknowledges with gratitude the funding support of Stanford University School of Medicine. icddr,b is also thankful to the following donors: the governments of Bangladesh, Canada, Sweden and UK for providing core/ unrestricted support. The donors had no role in data analysis, interpretation or decision to publish the findings.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.