Article Text
Abstract
Objectives Malaria is a vector-borne disease that remains a serious public health problem due to its climatic sensitivity. Accurate prediction of malaria re-emergence is very important in taking corresponding effective measures. This study aims to investigate the impact of climatic factors on the re-emergence of malaria in mainland China.
Design A modelling study.
Setting and participants Monthly malaria cases for four Plasmodium species (P. falciparum, P. malariae, P. vivax and other Plasmodium) and monthly climate data were collected for 31 provinces; malaria cases from 2004 to 2016 were obtained from the Chinese centre for disease control and prevention and climate parameters from China meteorological data service centre. We conducted analyses at the aggregate level, and there was no involvement of confidential information.
Primary and secondary outcome measures The long short-term memory sequence-to-sequence (LSTMSeq2Seq) deep neural network model was used to predict the re-emergence of malaria cases from 2004 to 2016, based on the influence of climatic factors. We trained and tested the extreme gradient boosting (XGBoost), gated recurrent unit, LSTM, LSTMSeq2Seq models using monthly malaria cases and corresponding meteorological data in 31 provinces of China. Then we compared the predictive performance of models using root mean squared error (RMSE) and mean absolute error evaluation measures.
Results The proposed LSTMSeq2Seq model reduced the mean RMSE of the predictions by 19.05% to 33.93%, 18.4% to 33.59%, 17.6% to 26.67% and 13.28% to 21.34%, for P. falciparum, P. vivax, P. malariae, and other plasmodia, respectively, as compared with other candidate models. The LSTMSeq2Seq model achieved an average prediction accuracy of 87.3%.
Conclusions The LSTMSeq2Seq model significantly improved the prediction of malaria re-emergence based on the influence of climatic factors. Therefore, the LSTMSeq2Seq model can be effectively applied in the malaria re-emergence prediction.
- infectious diseases
- public health
- information technology
- infection control
- epidemiology
Data availability statement
Data are available in a public, open access repository. Malaria cases for all 31 provinces of mainland China were obtained at https: www.phsciencedata.cn and the meteorological data at https://data.cma.cn/en.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
The use of climatic factors has proven to be effective predictors for malaria incidence and significantly affect the proposed long short-term memory sequence-to-sequence (LSTMSeq2Seq) model in capturing seasonal patterns and trends and predicting malaria incidence.
It is hard for a typical machine learning model to predict long-term dependencies, and it is even difficult for a single LSTM to capture key past events and use them to predict future values. By combining specialised LSTM cells that can forecast multiple time steps rather than having one multitasking cell, LSTMSeq2Seq solved this problem.
The LSTMSeq2Seq takes more time for training than other employed deep learning models. To train the LSTMSeq2Seq from scratch for all 31 provinces takes 2 weeks for four types of Plasmodium used in our study. Whereas other models take a few hours to days to train them using malaria cases and data of meteorological variables. In many provinces, LSTM was seven times faster than the LSTMSeq2Seq model. However, the impact is not significant in provinces with fewer malaria cases.
We could not obtain accurate predictions in some provinces by using any model in this study, due to the lack of other relevant potential non-climatic factors.
Introduction
Malaria is a vector-borne infectious disease caused by the parasitic protozoans of the genus Plasmodium such as Plasmodium falciparum (P. falciparum), Plasmodium ovale (P. ovale), Plasmodium vivax (P. vivax), Plasmodium simium (P. simium), Plasmodium knowlesi (P. knowlesi), and Plasmodium cynomolgi (P. cynomolgi). Governments, health organisations and scientific research institutions all over the world have made significant efforts on malaria control measures and elimination programmes. Despite the huge progress in reducing malaria cases and deaths, malaria remains life-threatening to global health mainly in Africa, Asia and America continents due to its sensitivity to environmental and climatic changes.
According to the World Malaria Report 2020 published by WHO, a total of 229 million malaria cases and 409 000 deaths were reported worldwide in 2019.1 Most of the malaria cases (93%) and malaria deaths (94%) occurred in the WHO African region, while the other WHO regions shared the remaining percentages.1 Despite remarkable progress, the global gains in fighting malaria disease have levelled off in recent years, and many high burdens have been losing ground. The combat against malaria had reached a crossroad.1 The world did not meet the milestones that could lower malaria cases and mortality by 90% by 2030. Without a massive coordinated action, the world is unlikely to meet the WHO’s Global Technical Strategy for malaria 2016–2030 targets.2 The COVID-19 pandemic has complicated the malaria picture even further, according to the WHO modelling analysis. The recent WHO report features a particular section on the COVID-19 pandemic and malaria, which could potentially double the number of malaria deaths in the WHO African region due to the disruptions to insecticide-treated net campaigns and the interruptions to access to antimalarial medicines.
Historically, malaria was one of the most prevalent parasitic diseases in the People’s Republic of China. However, through many years of combatting malaria, the Chinese government achieved remarkable progress in reducing malaria incidences through effective treatment and vector control measures. Vector control measures include reducing mosquito breeding grounds, implementing antimalaria grassroots campaigns.3 In 2010, the Chinese government launched the National Malaria Elimination Program.4–6 Indigenous malaria cases dramatically decreased to zero in 2017, which marked China among 21 countries with the potential of achieving a malaria eradication plan certified by WHO.7 However, imported P. falciparum malaria cases increased in many provinces, which poses a challenge to achieve malaria-free status and might cause another situation of malaria re-emergence that has been identified in some countries.8 9 A surveillance system in China is used to detect imported malaria cases but may miss some. Mosquitos are still out there with the ability to transmit the undetected imported malaria cases.
The re-emergence of malaria happened in Anhui and Henan provinces at the beginning of the 21st century. The re-emergence was due to climatic change, population movement, Anopheles abundance increase as well as mosquito’s drug resistance.10 11 Malaria outbreaks and re-emergence in the Huang-Huai River region happened due to the increase of Anopheles sinensis (An. sinensis). There was a high relationship between the re-emergence of P. vivax and an increase in the vectorial capacity of An. sinensis.12 13 Climatic conditions as the concerning factors in this study have contributed to the re-emergence of malaria by providing favourable conditions for the breeding and survival of mosquitoes.14 Numerous studies attempted to identify and assess the impact of climatic factors on malaria incidence in China.15–17 Some studies reported that the intra-annual variation in malaria cases might associate with changes in ambient temperature, precipitation, relative humidity, wind direction, sunshine duration and wind speed. Nevertheless, the findings were inconsistent in key factors observed and the corresponding effects estimated. Zinszer et al18 reviewed previously published studies related to the different approaches and factors used to predict malaria incidence. Most of the predictors were related to climate factors. Statistical, mathematical, machine learning and deep learning models have been applied to these climate predictors to improve the forecast accuracy of malaria incidence. Wang et al19 proposed an ensemble approach of traditional time series and deep learning models to improve the prediction performance of malaria incidence using malaria and climate data in Yunnan province. The study applied time series and deep learning models such as autoregressive integrated moving average (ARIMA), seasonal and trend decomposition using loess—integrated moving average (STL+ARIMA), backpropagation artificial neural network and long short-term memory (LSTM) network separately on the prepared data. Different evaluation methods were used to compare the prediction accuracy of methods. Gradient-boosting regression trees combine different models and are trained using climate data and malaria incidence. The model outperforms traditional time series and deep learning methods. Nkiruka et al20 proposed a machine learning system to assess the association between climatic factors and malaria incidence and found that rainfall, surface radiation and temperature affect the outbreak of malaria disease.
The relationship between malaria incidence and climatic factors is complex and cannot easily fit the classical forecasting approaches and machine learning algorithms. To reduce the complexity of this relationship by predicting malaria incidence with remarkable performance, deep learning models offer more advantages in the healthcare field by interacting with training data. Deep learning models give more accurate predictions compared with the statistical and mathematical approaches. Through deeper hidden layers, deep learning methods help us to gain unprecedented insights into care processes, diagnostics and forecasting and can make meaning from medical data. Deep learning models were applied to the prediction of directly transmitted infectious diseases.21 Some of the advanced deep learning models like LSTM and gated recurrent unit (GRU) recurrent neural networks with a large number of discrete time steps have been used in predicting infectious diseases like influenza, dengue incidence and hand, foot and mouth disease. LSTM model outperformed other machine learning models by achieving accuracy prediction and lower root mean squared error (RMSE).22–26 In this research, we identified and assessed climatic factors as predictors that may contribute to the re-emergence of malaria disease in China. We used climate factors with malaria incidence to train our constructed deep learning sequence-to-sequence model (LSTMSeq2Seq) and then evaluated its performance by predicting the re-emergence of malaria disease in China.
Methodology
Patient and public involvement
No patient involved.
Data collection and data preprocessing
We collected monthly malaria cases in all 31 provinces in China from January 2004 to December 2016. The data set contains four classes of Plasmodium species that is P. falciparum, P. vivax, P. malariae and other Plasmodium species. The plasmodium species category named other could be P.ovale, P. knowlesi or unidentified species type. Malaria cases for all 31 provinces of mainland China were obtained from the Chinese Center for Disease Control and Prevention (www.phsciencedata.cn)27 which provides the database for infectious diseases. The meteorological data of these 31 provinces were obtained from the China Meteorological data service centre (http://data.cma.cn/en).28 A total of 10 meteorological variables (ie, pressure, average temperature, maximum temperature, wind speed, minimum temperature, wind direction, precipitation, average relative humidity, sunshine duration, minimum relative humidity) were retained with no missing values in all features of meteorological data. To prevent overfitting while training the deep learning models, we used feature selection to remove redundant attributes. We reduced some of the meteorological variables using high correlation filtering and low variance filtering. Four variables (ie, pressure, wind speed, wind direction, sunshine duration) were discarded as they had the smallest variance in all the study areas. In total, 10 valid features (ie, six meteorological features and four types of malaria parasites) were considered in our study as shown in figure 1.
Guangdong climatic variables and P. falciparum used to train models. ARH, Average Relative Humidity; Avt, Average Temperature; MaxT, Maximum Temperature; MinT, Minimum Temperature; MRH, Minimum Relative Humidity; P. falciparum, Plasmodium falciparum.
Train-validation-test split
To train and evaluate the machine learning and neural network frameworks proposed in this paper, we divided the data set into the train, validation and test sets. In our experiment, 70% of the whole data set was used to train the model. We have allocated 15% of the data set for validation. The validation set was used to evaluate the model after each training epoch and ensure that the model is not overfitting the training data set. After the model has finished training, the remaining 15% of the data set was used to evaluate the model as the test set. The data was not shuffled before splitting to ensure that the validation set and test set results are more realistic. We allocated the period 1 January 2004 to 31 December 2012 to the training set and the period 1 January 2013 to 31 December 2014 is allocated to the validation set. The remaining period is allocated for the testing set.
Prediction models
This study proposes a sequence-to-sequence (Seq2Seq) prediction model based on the LSTM neural networks. The model will be used to forecast the re-emergence of malaria cases by considering the influence of meteorological factors on malaria cases in all 31 provinces of China. We compared the performance of our constructed LSTMSeq2Seq recurrent neural networks with other machine learning and deep neural networks prediction models, including XGBoost (extreme gradient boosting), GRU network and LSTM network models. Here is a brief description of our proposed Seq2Seq model as well as other employed models. These models achieved the best performance for predicting, diagnosing and controlling infectious diseases.
XGBoost model
The XGBoost is an ensemble machine learning algorithm that is flexible and easy to interpret. It provides an efficient implementation of gradient boosting machine learning model thought to be competent in the healthcare industry. A significant number of studies in public health have applied the XGBoost based framework to exploit data sources and predict infectious diseases such as dengue fever. The XGBoost model can achieve incredible performance in predicting vector-borne infectious diseases such as dengue or those caused by the West Nile virus.29 It has been used for forecasting, prevention and early diagnosis of infectious diseases30 31 and non-communicable diseases.32 The hyperparameters in this gradient boosting model were tuned to optimise the XGBoost model and achieve the best performance in our study. After testing several XGBoost parameters and the number of time steps as inputs, we chose 100 trees as the number of estimators to avoid overfitting. We used the GridSearchCV method in scikit-learn to tuning the hyperparameter and a learning rate of 0.8 and a maximum depth of 8. This method greatly reduces the prediction error of our XGBoost model. We used the defined types of monthly observation plasmodium incidence (P. falciparum, P. vivax, P. malariae and another class named other in our experiments) and climatic variables such as maximum temperature, average temperature, minimum temperature, average relative humidity, minimum relative humidity and rainfall to train the XGBoost approach and evaluate its performance on the test data set.
LSTM model
An LSTM describes a long short-term memory neural network and belongs to a class of recurrent neural networks (RNNs). RNN can process current data by using the previous data. It has effectively been used to solve problems of sequential time series such as climate modelling, web traffic prediction, financial prediction, neuroscience, intrusion detection, anomaly detection, air quality forecasting, medical monitoring. Meanwhile, RNN suffers from gradient vanishing and exploding problems when processing long-term dependencies sequences. The LSTM was developed as an intelligent recurrent neural network to specifically address the gradient vanishing problem by relying on memory cells, which have self-connections that store network temporal state, and are controlled by a set of three gates: input, output and forget. These gates and the memory cell can record information for a long time, thereby solving the problem of long-term dependencies and can predict the next time feature, which implies that it can forecast the next time step conditional on the previous values of the times series. LSTM’s ability to successfully learn from data with long-range temporal dependencies makes it a natural choice for time-series predictions. This model has achieved superior performance in predicting vector-borne infectious diseases like dengue fever33 and is one of the potential deep learning predictive models for childhood infectious diseases. It recently has been applied as one of the state-of-the-art deep neural networks in forecasting COVID-19.34–36 We developed a two-layer LSTM model that includes 128 and 32 memory cells and uses a batch size of 32 and a diagnostic of 1000 epochs. It consists of seven input parameters for each of the four classes of Plasmodium species, that is, P. falciparum. We have the monthly observation of P. falciparum incidences, maximum temperature, average temperature, minimum temperature, average relative humidity, minimum relative humidity and rainfall as the input vector sequence of the same month.
GRU model
GRU is an improved recurrent neural network as a simple variant of LSTM by combining the input gate and forgetting gate into a single gate called update gate. GRU comprises of update gate and resets gate, and it can only control information inside the unit because it has no additional memory cell to keep information. Researchers have applied this framework to forecast infectious diseases such as influenza.37 For the GRU model, we used the same hyperparameters as for LSTM models. The training data set was created using 12 months as input to our GRU model and the next month as output. The same input vector sequence as shown in figure 1 consists of seven input parameters for each of the four classes of Plasmodium species and six climatic variables. The six climatic variables, maximum temperature, average temperature, minimum temperature, average relative humidity, minimum relative humidity and rainfall, have been trained on the GRU model and used to test its performance.
LSTMSeq2Seq model
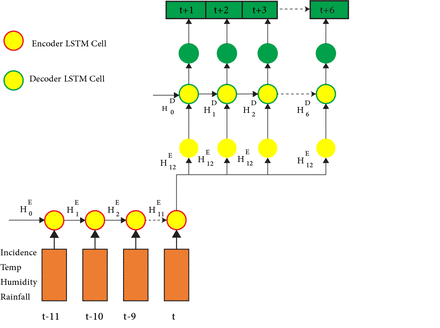
There are intuitively two different tasks to predict time series: understanding what has happened by looking at the known values of the past and predicting what will happen in the future. These two tasks require two different skill sets. The first is the ability to look at the past values and create an idea of the state of the system in the present. The second is the ability to use that understanding of the current state in the system to predict how the system will evolve in the future. As we mentioned earlier, LSTM predicts the next time feature, which implies that it can forecast the attribute of the next time step of input only. When we used a single LSTM cell in our model, we asked it to be capable of remembering both main events of the past and using those events to predict future values. Unlike single LSTM, we can use a Seq2Seq model with two specialised LSTM cells capable of predicting multiple time steps rather than having a single multitasking cell. Seq2Seq refers to the sequence-to-sequence architecture of the neural network fit. This architecture enables mapping between sequences of arbitrary length. As a result, Seq2Seq can perform many tasks, including language translation, image captioning and time series prediction. The Seq2Seq architecture is made up of an encoder and a decoder, as illustrated in figure 2.
Long short-term memory (LSTM) sequence-to-sequence architecture.
LSTMSeq2Seq model consists of two major blocks: encoder LSTM cell and decoder LSTM cell. The encoder outputs the encoder vector as input to the decoder block. The decoder encodes the input vector and predicts the next time step output. Subsequently, if Xt is the input of the next feature sequence, then the LSTM sequence model outputs Xtt+1 as the next time step feature.
The following are the formula for the encoder and decoder networks.
 (1)
(1)
where HEt represents the current hidden state at time step t, WHE is the appropriate weight of the old hidden state at time step t-1 and Wx represents the appropriate weight to the input vector Xt .
Equation (1) shows the result of a general sequence of the ordinary recurrent neural networks with the formula for the encoder. It is only necessary to apply an appropriate weight to the previous hidden state HEt−1 and the input vector Xt.
 (2)
(2)
where HDt is the current decoder hidden state, we are just using the old hidden state of the input vector at some time step t-1 to compute the next one and f is some function of the parameter.
Equation (2) is a stack of numerous recurrences that forecast each output yt at time t as a formula for the decoder. Each reiteration unit accepts a hidden state from the old unit and generates its hidden state.
The output yt at time step t is computed using the formula (3).
 (3)
(3)
yt is the final output state at time step t computed using softmax (is used to create a probability vector which will help us determine the final output) function and its respective weight Ws.
Equation (3) calculates the output using the state hidden at the current time step with each weight WS.
We designed an encoder that looks back into 12 months of historical data and a decoder that slide 6 months to predict, we have used t+12 months as input to the decoder as illustrated in figure 2 of our designed LSTMSeq2Seq model, the t+12 time step which is the encoder vector was used as input to the decoder and LSTM decoder cell predicts the next six steps ahead from t+1 to t+6 of malaria incidence. Apart from dropout, L1 regularisation and L2 regularisation were employed to avoid overfitting by preventing the weights of each network from being too high in the GRU, LSTM and LSTMSeq2Seq models. Each layer’s high parameter values can cause the network to concentrate severely on a few features, which can lead to overfitting. Weight regularisation added a cost to the loss function of the network for large weights. As a result, the models were forced to learn only the relevant patterns in the training data.
Model validation
Using two metrics loss function scores, we evaluated the performances of our methods for predicting the re-emergence of malaria incidence based on meteorological factors. First, we used RMSE as the basis for evaluating continuous variables by measuring the average differences between predicted and observed error values.
 (4)
(4)
where yt is the Plasmodium cases of observation for time t, and ŷt is the number of cases predicted by the model. A lower RMSE value indicates that there is a slight difference between the predicted Plasmodium cases and observed ones and implicates a high prediction accuracy of the model. Second, we used mean absolute error (MAE) to assess numerically the prediction error of the sequence and calculate the average value of the errors between Plasmodium cases of observation for the current time step and the predicted cases.
 (5)
(5)
Results
Comparison of LSTMSeq2Seq and candidate models
We performed all the experiments in Python (V.3.7.1) and modelled GRU, LSTM and LSTMSeq2Seq models through Tensor Flow (V.2.0.0), which is Google’s application programming interface for deep learning. We also used Keras (V.2.3.1), a deep learning library used in LSTM model development (Chollet, 2015).
The main goal of this study is to develop an accurate prediction model on the re-emergence of malaria cases based on the LSTMSeq2Seq neural networks using climatic factors and malaria incidence in 31 provinces of mainland China. We applied several machine learning and deep learning predictive models to achieve our goal. We evaluated the performance of four trained models: XGBoost, GRU, LSTM and LSTMSeq2Seq methods using the above evaluation metrics (RMSE and MAE). From tables 1–4, we show the RMSE/MAE of each model, with the LSTMSeq2Seq approach showing significantly lower errors than other approaches in almost all provinces and for all four species of Plasmodium malaria. The prediction errors have dropped significantly in many provinces as the LSTMSeq2Seq can improve the accuracy by learning the features and fluctuations of climatic variables on malaria incidence and predicting future cases. The following figure 3 illustrates the examples of the results predicted cases for P. falciparum, P. vivax, P. malariae and other based on the LSTMSeq2Seq prediction model. The Y-axis represents monthly number of malaria cases for each type of Plasmodium. The curves show that the peak value shifts downward for P. vivax as the time step predicted with accurate seasonal fluctuation compared with the P. falciparum. We selected the provinces presented in figure 3 based on two malaria high-risk zones according to the previous studies38 39: the central part of China along the Huai River that consists of Henan, Hubei, Anhui and Jiangsu provinces and the southwestern, southern regions which mainly comprising Guangdong, Guangxi, Hainan and Yunnan provinces. P. vivax was the dominant species in the first region as its climate is subtropical humid to subhumid monsoon. The LSTMSeq2Seq model achieved superior performance compared with other candidate models in most provinces with an average prediction accuracy of 87.3%. Models ranking from high performance to the lowest in the entire study are LSTMSeq2Seq, LSTM, GRU and XGBoost. LSTMSeq2Seq generates the lowest RMSE values of 0.0252, 0.0107, 0.0586 and 0.0077 for P. falciparum, P. vivax, P. malariae and other plasmodia, respectively. The LSTMSeq2Seq model reduced the mean RMSE of the predictions by 19.05% to 33.93%, 18.4% to 33.59%, 17.6% to 26.67% and by 13.28% to 21.34%, for P. falciparum, P. vivax, P. malariae and other plasmodia, respectively, as compared with other candidate models.
Comparison of model performances using the RMSE and MAE on the prediction of Plasmodium falciparum using climatic variables
Comparison of model performances using the RMSE and MAE on the prediction of Plasmodium vivax using climatic variables
Comparison of model performances using the RMSE and MAE on the prediction of Plasmodium malariae using climatic variables
Comparison of model performances using the root RMSE and MAE on the prediction of other Plasmodium species using climatic variables
Predicted cases for four Plasmodium types using long short-term memory sequence-to-sequence model.
Since 2008 the peak value shifted downward for P.vivax in different regions with a significant reduction but for the P. falciparum, there was an increase of trends which may be due to other factors apart from climate predictors like in Guangxi province in 2013 experienced the highest incidence because of the return of Chinese labours from gold mining in Ghana. However, the increasing trends of P. falciparum cases in Guangdong, Hainan and Jiangsu can be predicted well by LSTMSeq2Seq with superior accuracy to traditional machine learning model and better than deep learning state-of-the-art-models employed in this study. Thus LSTMSeq2Seq can be effectively applied to the prediction of malaria re-emergence in provinces with malaria incidence.
Discussion
In this study, we assessed the climatic factors that can affect the re-emergence of malaria incidence and built an advanced LSTMSeq2Seq deep neural networks model to predict the re-emergence of malaria in 31 provinces of China. We drew a comparison between the performance of the LSTMSeq2Seq model with other machine learning models applied in the study. The 2014 international panel report on climate change exposed an association between climate change and a significant increase in malaria burden.40 41 Previous studies suggested that climatic factors are not the only cause of malaria re-emergence since other non-climatic factors are also responsible.41 Besides climate change, malaria re-emergence is affected by other global changes such as demographic shifts, increased travel and trade. Although these non-climatic factors affect malaria transmission spatiotemporally, the climatic factors facilitate the transmission by providing a suitable environment for mosquito vector activities and Plasmodium incubation that cause an increase in the susceptible population. Based on these findings from the previous studies, we exploit the advantages of deep learning models in handling large data sets and use them to investigate the influence of climatic factors on malaria re-emergence. Researchers have developed malaria prediction models using climate determinants and malaria incidence data in different regions. However, to the best of our knowledge, this is the first time an LSTMSeq2Seq model was employed to construct a malaria re-emergence prediction model using climate determinants and malaria incidence data in all 31 provinces of China. By comparing the performance of the proposed model with that of other candidate models, LSTMSeq2Seq has proved to have a lower prediction error value in most of the provinces for different Plasmodium species. LSTMSeq2Seq has shown excellent ability to capture trends and seasonal patterns, especially for P. vivax and P. malariae, as most of the P. vivax cases were autochthonous and influenced by climatic factors, while P. falciparum cases may be imported and influenced by other global change factors. The climatic factors have proven to be effective predictors for malaria incidence and significantly affect the proposed LSTMSeq2Seq recurrent neural network models in capturing seasonal patterns and trends and predicting malaria incidence.
However, due to the fewer malaria cases in some provinces and a relatively small data set for a Seq2Seq deep neural network, GRU and XGBoost achieved lower RMSE/MAE values than the proposed method in some cases. Even so, the LSTMSeq2Seq model produced improved predictions and was better than other candidate models for each of the Plasmodium species in many provinces of China. However, for further improvement of malaria re-emergence prediction in China, our future research will consider climatic and non-climatic factors such as population movements, demographic shifts, changes in land use and civil unrest. By considering other potential factors that may contribute to the re-emergence of malaria incidence, we will increase the size of the data set and provide more patterns for Plasmodium species. We will also consider a deep learning technique known as transfer learning. This technique uses the learnt tusk related to the new tusk to accelerate its training and improve its predictive accuracy. It will reduce the prediction error value of the LSTMSeq2Seq in the provinces with fewer malaria cases through transfer from the previously trained model in regions with high malaria cases. Based on the LSTMSeq2Seq model, this research achieved accurate prediction of malaria cases in China, using long-term time series malaria cases and the data of climatic variables. This method might be used for the large-scale prediction of other malaria-like diseases.
There are some limitations to this study. First, the LSTMSeq2Seq takes more time for training than other employed deep learning models. To train the LSTMSeq2Seq from scratch for all 31 provinces takes 2 weeks for four types of Plasmodium used in our study, whereas other models take a few hours to days to train them using malaria cases and data of meteorological variables. For most cases, LSTM was seven times faster than the LSTMSeq2Seq model. However, the impact model is not significant in provinces with fewer malaria cases. Second, we could not obtain accurate predictions in some provinces by using any model in this study, probably because we failed to get other relevant potential non-climatic factors.
Conclusion
Malaria is still a public health burden that can be widely transmitted through the influence of many factors. To reduce this burden, it is very important to predict the re-emergence of malaria and put in place serious control measures. In this study, we investigated the influence of climatic factors in the re-emergence of malaria in mainland China by proposing an LSTMSeq2Seq model capable of effectively predicting malaria incidence using climatic factors and different types of Plasmodium species in all 31 provinces of China. We compared typical machine learning and other recurrent neural networks models with the performance of the LSTMSeq2Seq approach. Remarkably, the prediction performance observed in this paper indicates that LSTMSeq2Seq prediction performance outperforms the other candidate models applied in the study. Therefore, the LSTMSeq2Seq model can be effectively applied in the malaria re-emergence prediction.
Data availability statement
Data are available in a public, open access repository. Malaria cases for all 31 provinces of mainland China were obtained at https: www.phsciencedata.cn and the meteorological data at https://data.cma.cn/en.
Ethics statements
Patient consent for publication
Ethics approval
The research protocol was approved by the institutional review board of the Institute of Complexity Science, Qingdao University, China.
References
Footnotes
Twitter @kameri16
Contributors EK analysed and preprocessed the data, trained and evaluated the performance of the models, interpreted results and wrote the manuscript. JZ supervised, coordinated the design of the entire study and reviewed and edited the manuscript. JZ was the guarantor. DB collected the data used in this study. All authors have read and agreed to the submission version of the manuscript.
Funding This work was supported by the Shandong Provincial Natural Science Foundation, China (ZR2018MH037).
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.