Article Text
Abstract
Introduction The intestinal microbiome in early life plays a major role in infant health and development. Factors like antibiotic exposure, breast/formula feeding and mode of delivery are known to affect the microbiome. The increasing occurrence of caesarean section (C-section) deliveries and antibiotic exposure warrants further insight into the potential missing microbes in those infants. The study objective is to study the effect of maternal antibiotic administration during pregnancy and/or C-section mode of delivery on the development of the infant’s intestinal microbiome until the age of 2 years.
Methods and analysis A single site, cross-sectional observational study of C-section and vaginally delivered infants being either exposed to maternal antibiotic treatment or not during the third trimester of pregnancy. Throughout the nine visits, stool, urine, saliva, hair, breast milk and vaginal swabs will be collected from either mother and/or infant for microbiome and metabolomic analysis.
Ethics and dissemination The protocol was approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals. The trial has been registered at ClinicalTrials.gov.
The findings from this study will be disseminated in peer-reviewed journals, during scientific conferences, and directly to the study participants. Sequencing data will be deposited in public databases.
Trial registration number NCT04134819.
- MICROBIOLOGY
- PAEDIATRICS
- OBSTETRICS
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
Methods that allow to study the effects of maternal antibiotic administration during pregnancy and/or caesarean section (C-section) mode of delivery on the development of the infant’s intestinal microbiome until the age of 2 years.
Methods described in this study will allow for identification of the missing microbes in babies due to C-section delivery/antibiotic exposure, which is the first step towards replenishing the depleted microbiome and possibly limiting the negative impact of C-section delivery/antibiotic exposure on the infant’s microbiome, metabolome and development.
The study design includes a longitudinal aspect allowing for monitoring the development of an infant’s gut microbiome during the first 2 years of life.
This is a single-site study.
Introduction
The intestinal microbiome in early life plays a major role in infant health and development, impacting the maturation of the immune system, protection against pathogens and influencing the long-term metabolic welfare of the host.1 The acquisition of microbial strains from mother to infant may occur through multiple different pathways, including the birth canal (and the proximity of the birth canal to the anus), contact between mothers and infant during parental care and through breast milk.2 Breast milk is a natural prebiotic source that provides the optimal active ingredients for the growth of beneficial microbial species in the infant intestine. The microbiome of vaginally born, exclusively breastfed infants at term, with no previous exposure to antibiotics either directly or indirectly from the mother, could be considered the ‘golden standard’. In contrast, antibiotic exposure, caesarean section (C-section) birth or formula feeding are known to be associated with altered gut microbiome compared with ‘golden standard’ babies, and this may be associated with poorer health status.3
Antibiotic treatment throughout pregnancy accounts for up to 80% of prescribed medications during pregnancy.3 It is estimated that one in five pregnant women in Europe is prescribed at least one course of antibiotics during pregnancy.4 Considering that it is now accepted that an optimal establishment of the gut microbiome in infants is highly desirable for the normal development of a human, the risk is that antibiotic usage during pregnancy may have undesirable effects on the maternal vaginal, gut and milk microbiome, with knock-on negative impact on the early infant gut microbiome.
The gut microbiome and its perturbations appear to also influence the development of emotional behaviour, stress and pain modulation systems and brain neurotransmitter systems.5 A lesser performance in neurodevelopmental outcomes and cognitive development was demonstrated in C-section-delivered infants6 and children7 compared with vaginally delivered ones. A meta-analysis revealed that C-section delivery was significantly positively associated with autism spectrum disorder and attention-deficit/hyperactivity disorder in infants.8 Potential developmental delay in the children is assessed with the Bayley Scales of Infant and Toddler Development Version III (BSID-III).9 The BSID-III assesses three areas of development by trained healthcare professionals using: cognitive scales, receptive and expressive language scales, as well as fine and gross motor scales. Questionnaires are completed by the parents and focus on the social-emotional scale and adaptive behaviour scale.9
The development of the infants’ microbiome may be affected by maternal depression during pregnancy and postnatally, and is a condition affecting up to 17% of mothers and indirectly their infants.10 The Edinburgh Postnatal Depression Scale (EPDS)11 is a simple 10-point Mental Health Questionnaire that assesses if the mother has symptoms that are common in women with depression and anxiety.
This project will investigate the impact of antibiotic treatment during pregnancy on the vaginal, intestinal and milk microbiome of the pregnant mother and antibiotic exposure and C-section delivery on the intestinal microbiome composition and functionality of her offspring during the first 2 years. The study intends to reveal which populations of microbes are most affected by these perinatal factors. The study will also investigate the health readouts including anthropometric measurements, infant development and salivary cortisol as stress marker. The study will create the basis for the development and testing of early life nutritional interventions, focusing on ‘next generation’ microbial therapeutics.
Methods
Study design
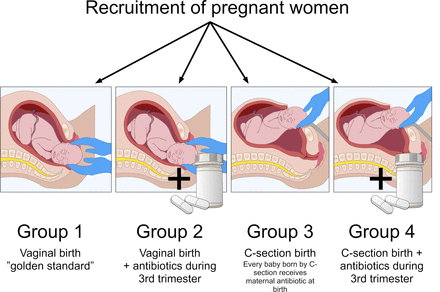
This study is a cross-sectional observational study of pregnant women and their C-section and vaginally delivered infants being either exposed to maternal antibiotic treatment during the third trimester of pregnancy or non-exposed (figure 1). This single-site study will investigate the differences in the microbiome and metabolomics readouts and stress levels between exposed and non-exposed participants and mode of birth in a newborn population.
Study design, representing four comparative arms and the approximate number of infants per group (group 1 - 168, group 2 - 112, group 3 - 72, group 4 - 48) assuming that the probability for a mother to receive antibiotics during pregnancy is the same for the two delivery modes. Nonetheless, note that every baby born by C-section indirectly receives antibiotics at birth. Copyright Pinja Kettunen/SciArt & IFF Health & Biosciences, with permission. C-section, caesarean section.
The study started in August 2018 and is planned to finish by March 2023.
Participant selection
The participants are pregnant females of age 22–40 years, who wish to breastfeed their infants and deliver their singleton pregnancies at greater than 35 weeks’ gestation at the Cork University Maternity Hospital, Ireland. The study aims to enrol a total of 500 participants to have 400 mother–infant pairs completing the study, due to an expected overall 20% drop-off rate (based on the pilot study and previous experience).
Approximately 40% of the pregnant women are anticipated to receive antibiotics during the third trimester of the pregnancy, mainly due to urinary tract infections12 and upper and lower respiratory infections.13 Therefore, the total cohort of 400 women completing the study will include non-antibiotic controls (approx. n=240) and antibiotic-treated women (approx. n=160). Approximately 30% of infants will be born by C-section (approx. n=120), while 70% will be born vaginally (approx. n=280)14 (figure 1).
Inclusion and exclusion criteria
To be eligible for the study, the participants must meet the terms of the inclusion and exclusion criteria as presented in table 1.
Inclusion and exclusion criteria of the missing microbes in infants born by Caesarean section (MiMIC) study
Recruitment
Pregnant women will be approached during the third trimester at the antenatal clinics and an arrangement will be made to meet with them on their next hospital visit to obtain consent and request a stool sample. Study-related information will be given in written form as well as explained by the research nurse. No study-related activities will begin before the potentially eligible participants have signed the informed consent form (ICF; online supplemental file 1). Participants will also receive and sign a data protection notice.
Supplemental material
Compensation
There is no compensation provided to the participants. There are no cost implications for the Health Service Executive (HSE) or to the participants. The management of patients and investigative tests will comply with current standards of care.
Study timeline
In total, the study will include nine visits during the first 2 years (figure 2). Throughout the study, stool, urine, saliva, hair, breast milk and vaginal swabs will be collected from either mother and/or infant. All visits will be the same for both C-section and vaginally delivered infants. The mothers will be provided instructions for sample collection as well as required materials during the visits. During each visit, the antibiotic exposure and medical status of the mother and infant will be recorded based on the medical chart and/or mother’s report in the case report form (CRF).
Visit schedule, biological sample and data collection points during the study (ClinicalTrials.gov, NCT04134819). Copyright Pinja Kettunen/SciArt & IFF Health & Biosciences, with permission.
Study visit 1
During the screening visit, after completing recruitment procedures, that is, reading and signing the ICF and reviewing the inclusion and exclusion criteria, demographic data will be collected from the pregnant woman in the CRF including her date of birth, occupation, education, presence of pets in the household, parity, gestation, delivery due date and smoking status (smoker/non-smoker/former smoker/never smoker). A stool sample will be requested from the mother. She will be asked to complete a Mental Health Questionnaire (Edinburgh Postnatal Depression Scale; EPDS11) and a Food Frequency Questionnaire (FFQ15). Maternal weight and height will be obtained. This may take two meetings with the participant to complete.
Study visit 2
At birth, a vaginal swab will be obtained from women undergoing vaginal delivery. The infants’ date of birth, sex and mode of delivery will be recorded.
Study visit 3
One-week postpartum, a breast milk sample will be collected from the mother, while stool and hair samples will be collected from the infant.
Study visit 4
Four weeks postpartum, the mother will provide a stool sample, a breast milk sample and a hair sample. Stool and urine samples will be collected from the infant.
Study visit 5
Eight weeks postpartum, breast milk and saliva samples will be collected from the mother. The mother will also complete the EPDS Mental Health Questionnaire, while stool and saliva samples will be collected from the infant.
Study visit 6
At 6 months of age, a stool sample will be collected from the infant. A breast milk sample will be collected if the mother is still breast feeding. The mother will also complete a FFQ.
Study visit 7
At 1 year of age, a stool sample will be collected from the infant. A breast milk sample will be collected if the mother is still breast feeding.
Study visit 8
When the infant is 18 months old, a stool sample will be collected from the infant, while a breast milk sample will be collected from the mother if she is still breast feeding.
Study visit 9
At the age of 2 years, a stool sample will be collected from the infant. In addition, the Bayley Scales assessment of the infant will be performed (Bayley Scales of Infant and Toddler Development Version III; BSID-III9). If the mother is still breast feeding, a breast milk sample will be collected.
Participant withdrawal/exclusion
Under the Declaration of Helsinki, the research nurse will explain to the participant that they have the right to withdraw from the study at any time and that this will in no way prejudice their future treatment. The reason for withdrawal will be recorded in the source documents and on the appropriate CRF. Withdrawn participants will be replaced. Samples from individuals withdrawing from the study participation will be stored and may be analysed in the study. Participants excluded from the study after consenting will be replaced.
Ethics and dissemination
The study is conducted following the version Fortaleza, Brazil, October 2013 of the Declaration of Helsinki 1964. The Protocol and the ICF have been approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals (CREC) before commencement (approval letter ECM 4 (q) 07/03/18 & ECM 3 (ppppppppp) 10/04/18). On 17 February 2022, Protocol version 12 has been approved by the CREC (approval letter ECM 4 (q), 7 March 2018 and ECM 3 (ttttt), 22 February 2022).
If a protocol amendment is necessary, this will be prepared with the agreement of the chief investigator and signed by the relevant parties. If the amendment is substantial, it will be submitted to the CREC and possibly other public bodies according to local requirements for review and approval. The protocol amendment will not be implemented before the required approvals are obtained.
The trial is sponsored by University College Cork (College Road, Cork T12 K8AF, +353 (0)21 490 3000). The sponsor is involved in study design; collection, management, analysis and interpretation of data; writing of the report; and the decision to submit for publication. The funder was involved in the study design; analysis and interpretation of data; writing of the report; and the decision to submit for publication. The trial has been registered at ClinicalTrials.gov.
The findings from this study will be disseminated in peer-reviewed journals, during scientific conferences and directly to the study participants.
Objectives and outcomes
Primary objectives and outcomes
To study the effect of maternal antibiotic administration during pregnancy and/or C-section mode of delivery on the development of the infant’s intestinal microbiome until the age of 2 years.
To select a cohort of vaginally delivered infants to isolate the ‘missing microbes’ in C-section and/or antenatally antibiotic exposed infants.
Secondary objectives and outcomes
The effect of maternal antibiotics on the developing infant as evaluated by:
Anthropometric assessment:
Change in infant’s body weight from birth to 6, 12 and 24 months
Change in infant’s body length from birth to 6, 12 and 24 months
Change in infant’s head circumference from birth to 6, 12 and 24 months.
Bayley Scales of Infant and Toddler Development test at 2 years of age.
Ancillary objectives and outcomes
To isolate and characterise bacterial strains from fresh healthy infant faeces that are altered in the stools from C-section delivered and antibiotic-treated infants compared with vaginally delivered infants.
The effect of maternal antibiotic treatment on the human milk microbiome during lactation.
The effect of maternal antibiotic treatment on the mother’s vaginal microbiome at birth.
The effect of maternal antibiotic treatment on the metabolomics of infant’s urine and stool at 4 weeks of age.
Stress hormone (cortisol) levels in saliva and hair of mothers and infants up to 8 weeks postpartum.
EPDS Mental Health Questionnaire of mothers at third trimester and 8 weeks postpartum.
FFQ of mothers at third trimester and 6 months postpartum.
Changes in maternal microbiome from pregnancy until 4 weeks postpartum.
Exploratory objectives and outcomes
To identify, isolate and characterise bacterial strains that can be further developed into probiotic products to help replenish depleted microbiome in the gut of infants born by C-section and/or by antibiotic-treated women.
Sample collection and analysis
Faecal samples
Faecal samples will be collected from mothers and infants. Mothers will collect a whole bowel motion from themselves in a stool container and mothers will also collect as much as possible of the infant’s faeces (at least three to four scoops; 0.2 g) from the infant’s nappy into a stool container throughout the study. Mothers are instructed to place the filled stool containers into a red bag and place it into a cool place. Within 10 hours (typically 4–5 hours) after being collected, a research nurse will transfer samples in coolers to the laboratory where the samples may be split for DNA extraction, metabolomics and microorganism culturing. In the first two cases, samples will be stored at −80°C until further analysis; in the latter case, sample will be kept anaerobically at 4°C until plating on up to 14 different culture media (four non-selective (brain heart infusion agar (BHI); tryptic soy agar (TSA); yeast, casitone and fatty acid agar (YCFA); gut microbiota medium agar (GMM)) and 10 species-specific selective media (supplemented brain heart infusion agar (BHIS); Bifidobacterium selective agar; Lactobacillus selective agar; De Man, Rogosa and Sharpe agar; Reinforced clostridial agar; MacConkey; Fastidious anaerobic agar with horse blood; Tryptic soy yeast extract agar; Kanamycin esculin azide agar; Colombia agar)] to cover the widest possible range of the gut microbiota. DNA will be extracted from stool using the modification of the Repeated Bead Beating Plus Column (RBB+C) method16; (online supplemental file 2) and subjected to 16S rRNA and/or shotgun sequencing. Metabolomics of infant’s faeces will be analysed by the gas chromatography–mass spectrometry (GC-MS) technique using methyl chloroformate derivatisation.17
The bioinformatic analysis will identify the microbial composition at the phylum, genus and family levels using both read-based18 and assembly-based19 20 approaches. Metagenomic data will be correlated with physiological and clinical parameters and biomarkers. Advanced statistical and bioinformatics approaches will be applied to identify modifiable environmental factors that influence bacterial groups/consortia as well as immune and brain development and function. This includes diversity analysis,21 differential abundance analysis,22 strain persistence and vertical transmission23 and genome-scale metabolic modelling.24
Milk samples
Breast milk samples will be collected from mothers throughout the study as long as they are breast feeding and have an adequate milk supply to give samples for research purposes. Mothers will be instructed that once their infant is satisfied, to clean their breast with provided sterile water swab and to express a sample of breast milk (10 mL) and place it into the refrigerator (4°C). The sample will be collected by the research nurse within 10 hours (typically 4–5 hours) after being expressed by the mother and transported in a cooler to the laboratory. The samples may be split for DNA extraction (using modification of Qiagen DNeasy Powerfood Microbial Kit (QIAGEN Ltd, Manchester, UK; online supplemental file 3) and microorganism culturing. In the first case, samples will be stored at −80°C until further analysis; in the latter case, sample will be kept anaerobically at 4°C until processing. The milk microbiome composition will be analysed by using state-of-the-art methods, possibly including but not limited to sequencing, and quantitative PCR.
Urine samples
A urine sample (5 mL) will be obtained from the infant 4 weeks postpartum in the morning. This will be collected by placing a sterile pad in the infant’s nappy and removing the pad when next changing the nappy. Sample to be stored at 4°C until pick-up by the research nurse. The urine samples will be transported in a cooler to the laboratory, where urine will be recovered and stored at −80°C. Urine metabolomics analysis will be analysed by the Liquid chromatography–mass spectrometry (LC-MS) technique.25
Hair samples
A hair sample will be obtained from the infant 1 week postpartum and from the mother at 4 weeks after birth. The sample will be obtained from the base of the cranium. If the infant has adequate hair, a 1 cm long sample will be collected and placed in a ziplock bag. From the mother, 20–30 strands of 3 cm long hair (tied with an elastic band) will be cut close to the scalp and wrapped in tin foil and placed in a ziplock bag. Hair samples will be stored in the laboratory at 4°C. Hair cortisol analysis is to be carried out by extracting cortisol through a combined process of methanol extraction and sonication after which the samples are evaporated to dryness using nitrogen gas (using modification of the Kalra et al method26; online supplemental file 4). Cortisol levels will be measured using a commercially available ELISA kit (Cortisol ELISA kit, Enzo, USA).
Saliva samples
A liquid saliva sample will be collected 8 weeks after birth using sterile SalivaBio’s Children’s Swab (Salimetrics, USA) and SalivaBio Infant Swab (Salimetrics) from the mother and the infant, respectively. The liquid samples retrieved from the swab by centrifugation (2683 g for 10 min) will be stored at the laboratory at −80°C until the measurement of salivary cortisol levels using the commercial kit (Cortisol ELISA kit, Enzo).
Vaginal swabs
A midlumen swab will be collected for vaginal deliveries from the mother by the attending midwife in the delivery suite. Once collected, the sample will be stored at 4°C in the delivery suite. Collected swab samples will be transported to the laboratory and stored at −80°C until processing. DNA will be extracted from vaginal swabs using the modification of the RBB+C method16 (online supplemental file 2). The vaginal microbiome composition will be analysed by using state-of-the-art methods, possibly including but not limited to sequencing, and quantitative PCR. The bioinformatic analysis will identify the microbial composition at the phylum, genus and family levels.
Adverse events and participant well-being
There are no expected safety concerns related to the study. All self-reported adverse events will be listed documenting duration, severity, subject outcome and if any therapy was required. The study chief investigator will review these and advance any reports to local ethics committee if it is deemed necessary. In case a mother completing the EPDS questionnaire scored above the screener’s threshold for risk of antenatal depression, participant will be contacted by the research nurse. If a participant feels that she might benefit from additional support and consent to sharing her high EPDS score with her general practitioner (GP), a letter will be sent to her GP and appropriate referrals made.
Data collection and management
The study diaries, questionnaires and paper/digital CRF systems will be used for recording data from each participant meeting the eligibility criteria and being included in the study. During the study, the paper-based system will be updated to the Castor electronic data capture platform (www.castoredc.com/). All the captured data in this trial are pseudonymised, as each of the participants will receive a specific study code, and on receipt, data will be referring to the study code. All study staff responsible for entering data into the CRF system will be trained before commencing their work. A minimum of four monitoring activities will be performed: site initiation visit, two visits during the trial and a close-out visit; this will be done according to the monitoring plan. For details, see online supplemental file 5. Any inconsistencies identified during the study will be presented as queries.
Data types and formats collected and/or produced will include raw whole genome sequencing (WGS) data (fastq), WGS assembly files and Excel file with strains name (fasta), WGS annotation files (fna, gff, faa, ffn), 16S raw data (fastq), 16S ASV table (csv), 16S ‘phyloseq’ file, which includes metadata for each sample (rds), Raw shotgun data (fastq), metagenome-assembled genomes (MAGs)/bins from shotgun data (fna), pangenomes files (newick, fa, csv/tab) and pipeline for the metabolic modelling part and generated data (sbml, mat). Sequencing data will be deposited in public databases.
Statistical analysis
Sample size justification
In microbiome studies, power analyses can be difficult because the true effect size is unknown and also because the composition of the microbiome in control and experimental groups (ie, beta-diversity) is generally unknown. However, in this study, the aim is to use pairwise distances and permutational multivariate analysis of variance (PERMANOVA) to measure diversity based on taxonomic data, and so we can use simulation to estimate statistical power.27 In short, power is calculated from PERMANOVA tests by estimating an effect size based on datasets characteristic of one of the groups, here the ‘golden standard’ group (group 1). Representative data were downloaded from the INFANTMET study (ie, stool samples from 4-week-old infants).25 Then, distance matrices (here the weighted Jaccard distance) are simulated with the same properties as the original dataset, and power is estimated by bootstrapping the simulated distance matrices.
To detect a small effect size of 0.0206 (p value <0.001) with >90% power would require 82 individuals in the ‘golden standard’ group (and assuming the same number in the three other groups). This number of individuals is very close to similar studies (see, eg, Lundgren et al28). Here, the size of the different groups will not be equal (figure 1). For group 3 and Group 4, we obtain a power of 87% and 71%, respectively. However, the effect size calculated from the ‘golden-standard’ group is very small, and assuming a slight increase of the effect size to 0.03, we obtain a power >90% for these two groups. Additionally, we will perform shotgun metagenomics analysis on 25% of all the samples, a technique that can reach strain-level precision as well as a better precision for low-abundant genera than 16S rRNA amplicon sequencing.29 This ultimately refines the calculation of diversity and allows for better discrimination between samples as well as increases the observed size effect. By coupling 16S rRNA and shotgun metagenomics, we thus assume that the study will be powered for all the groups. We will calculate and mention post hoc power analysis to confirm it.
Patient and public involvement
None.
Discussion
Throughout the MiMIC study, stool, urine, saliva, hair, breast milk and vaginal swabs will be collected from either mother and/or infant. Collected samples will allow us to study the effect of maternal antibiotic administration during pregnancy and birth mode on the development of the infant’s intestinal microbiome until the age of 2 years. The impact of maternal antibiotic treatment and/or C-section birth mode on the infant gut microbiome will be also analysed through differences in metabolomics, as changes in the microbiome are likely to be reflected in changes in microbially derived metabolites. Eventually, we will collect data sets to evaluate the effects of maternal antibiotic treatment and/or birth mode on infant’s development using the BSID-III. External factors such as maternal diet, stress levels, mental status and medication have the potential to affect the microbiome; thus, with the data collected in the MiMIC study, we will be able to minimise the effects of those factors. Identification of the missing microbes, microbes missing in babies due to C-section delivery/maternal antibiotic exposure, is the first step towards replenishing the depleted microbiome and possibly limiting the negative impact of C-section delivery/maternal antibiotic exposure on the infant’s microbiome, metabolome and development. If successful, those identified, isolated and characterised missing microbes can be further developed into ‘next generation’ microbial therapeutics to help restore microbiome that has been depleted in the gut of infants born by C-section and/or by antibiotic-treated mothers.
Impact of COVID-19
COVID-19 pandemic emerged in March 2020 as a major disruption in primary care clinics. At the time, 282 participants out of 500 had been recruited, with recruitment expected to be completed originally in June 2021 (figure 3). Recruitment outside of the hospital setting was not possible until 20 July 2020, while recruitment in the hospital setting could only resume from 5 October 2020. In 2021, recruitment was paused again from 4 January 2021 to 15 April 2021, when online recruitment via social media could commence. A steady increase in the number of recruited participants has been observed since then, and currently (29 Septembre 2022), 485 participants have consented corresponding to 97% of the study target, and we estimate recruitment completion during Q1 2023.
Effect of COVID-19 on participant recruitment showing the targeted (grey) and actual recruitment (black) rate.
Interestingly, during the COVID-19 pandemic, we observed a lower than expected occurrence of antibiotic exposure during the third trimester of pregnancy among study participants. To assure sufficient power to all the study groups, since July 2021, we are also approaching women in the delivery unit that are known to be exposed to antibiotics in the third trimester. The known limitation of this approach is the omission of the mother stool sample during third trimester. Nevertheless, this approach in combination with continued recruitment of pregnant women is the valid strategy to increase the number of participants exposed to antibiotics in the third trimester (groups 2 and 4).
Due to the governmental restrictions during the COVID-19 pandemic, sample collection was paused from March 2020 to 27 May 2020. The initial study design assumed a 1-week window for collection of the samples at 6, 12, 18 and 24 months. To comply with the governmental requirements and to maximise outcomes from active participants, we increased the sample collection window by 3 months. The frequency of sample collection at earlier time points could not allow for the expansion of the sample collection window. This adjustment due to COVID-19, resulted in loss of only 20 mother stool samples, 69 breast milk and 69 infant stool samples. These samples represent less than 5% of the total number of samples collected at a given time point. At the same time, we have introduced several adjustments to the sample collection; we suspended the brief physical examination as well as anthropometric measurements of the infant during the sample collection; the dispensing of sample containers is now done during the collections to eliminate unnecessary interactions; the vaginal swabs are not collected until the hospital policies allow.
Despite the COVID-19 pandemic, we maintained high participant retention, with over 95.4% of consented participants continuing in the study (including 94 excluded participants), and as of 29 September 2022, 190 participants have reached visit 9.
Status of trial
The trial is ongoing, and as of 29 September 2022, 97% of the participants have been recruited.
Ethics statements
Patient consent for publication
Acknowledgments
The authors would like to thank all the study participants.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Contributors AKW wrote the manuscript. EMD, SDF, CAR, JFC, EP, MNO’R, C-AO'S, RPR and CS were involved in the study design and writing of the manuscript. C-AO’S, FK, GM and OO’C were involved in consenting participants and collecting samples. All authors read and approved the final manuscript.
Funding This publication has emanated from research conducted with financial support of Science Foundation Ireland under Grant No. 12/RC/2273_P2 and 19/SP/6989. This publication has emanated from research conducted with financial support of International Flavors & Fragrances (IFF).
Competing interests JFC has been an invited speaker at meetings organised by Mead Johnson, Yakult, Nutricia, Pepsi and Friesland Campina and has received research funding from Cremo, IFF, Pharmavite, Mead Johnson and Nutricia; and has been a consultant for Nestle. SDF is an employee of IFF Health & Biosciences that manufactures and commercialises probiotics, while EP was an employee of IFF Health & Biosciences at the time this manuscript was prepared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.