Article Text
Abstract
Objectives Lung ultrasound (LUS) is a portable, low-cost respiratory imaging tool but is challenged by user dependence and lack of diagnostic specificity. It is unknown whether the advantages of LUS implementation could be paired with deep learning (DL) techniques to match or exceed human-level, diagnostic specificity among similar appearing, pathological LUS images.
Design A convolutional neural network (CNN) was trained on LUS images with B lines of different aetiologies. CNN diagnostic performance, as validated using a 10% data holdback set, was compared with surveyed LUS-competent physicians.
Setting Two tertiary Canadian hospitals.
Participants 612 LUS videos (121 381 frames) of B lines from 243 distinct patients with either (1) COVID-19 (COVID), non-COVID acute respiratory distress syndrome (NCOVID) or (3) hydrostatic pulmonary edema (HPE).
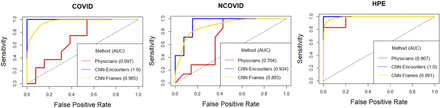
Results The trained CNN performance on the independent dataset showed an ability to discriminate between COVID (area under the receiver operating characteristic curve (AUC) 1.0), NCOVID (AUC 0.934) and HPE (AUC 1.0) pathologies. This was significantly better than physician ability (AUCs of 0.697, 0.704, 0.967 for the COVID, NCOVID and HPE classes, respectively), p<0.01.
Conclusions A DL model can distinguish similar appearing LUS pathology, including COVID-19, that cannot be distinguished by humans. The performance gap between humans and the model suggests that subvisible biomarkers within ultrasound images could exist and multicentre research is merited.
- chest imaging
- ultrasound
- respiratory infections
- adult intensive & critical care
- COVID-19
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
The ability of a convolutional neural network (CNN) to differentiate between similar appearing lung ultrasound (LUS) images with pathological B lines of three different origins (COVID-19, non-COVID acute respiratory distress syndrome and hydrostatic pulmonary edema) was evaluated using 612 LUS videos from 243 patients.
The performance of the neural network was benchmarked against physicians competent in LUS interpretation who completed an online interpretation exercise.
The performance of the neural network was evaluated using a batch of 10% of our data that was held back from the training process, thereby estimating generalised performance and defending against model overfitting that may otherwise embellish deep learning (DL) results.
Explainability efforts using heatmap-based tools were employed to allow insight into the regions of LUS images that contributed most to the predictions of the neural network.
Our study used the largest volume of data to date for a DL problem related to LUS, although this amount of data is small and more heterogeneous relative to other imaging-based DL models.
Introduction
Lung ultrasound (LUS) is an imaging technique deployed by clinicians at the point-of-care to aid in the diagnosis and management of acute respiratory failure. With accuracy matching or exceeding chest X-ray (CXR) for most acute respiratory illnesses,1–3 LUS additionally lacks the radiation and laborious workflow of CT. Further, as a low-cost, battery-operated modality, LUS can be delivered at large scale in any environment and is ideally suited for pandemic conditions.4
B lines are the characteristic pathological feature on LUS, created by either pulmonary edema or non-cardiac causes of interstitial syndromes. The latter includes a broad list of conditions ranging from pneumonia, pneumonitis, acute respiratory distress syndrome (ARDS) or fibrosis.5 While an accompanying thick pleural line is helpful in differentiating cardiogenic from non-cardiogenic causes of B lines,6 reliable methods to differentiate non-cardiogenic causes from one another on LUS have not been established. Additionally, user-dependent interpretation of LUS contributes to wide variation in disease classification,7 8 creating urgency for techniques that improve diagnostic precision and reducing user dependence.
Deep learning (DL), a foundational strategy within present-day artificial intelligence techniques, has been shown to meet or exceed clinician performance across most visual fields of medicine.9–11 Without cognitive bias or reliance on spatial relationships between pixels, DL ingests images as numeric sequences and evaluates quantitative patterns that may reveal information that is unavailable to human analysis.12 With CT and CXR research maturing,13–15 LUS remains comparably understudied with DL due to a paucity of organised, well-labelled LUS datasets and the seeming lack of rich information in its minimalistic, artefact-based images.
In this study, we trained a neural network using LUS images of B lines from three different aetiologies (hydrostatic pulmonary edema, ARDS and COVID-19). Using LUS-fluent physicians as comparison, we sought to determine if subvisible features in LUS images are available to a DL model that would allow it to exceed human limits of interpretation.
Methods
Data identification, extraction and labelling
After University of Western Ontario Research Ethics Board (REB 115723) approval, LUS examinations performed at London Health Sciences Centre’s two tertiary hospitals were identified within our database of over 100 000 point-of-care ultrasound examinations. The curation and oversight of this archive have previously been described.16 The goal of this study was to determine if a deep neural network could distinguish between the B line profiles of three different disease profiles, namely, (1) hydrostatic pulmonary edema (HPE); (2) non-COVID ARDS (NCOVID) causes; and (3) COVID-19 ARDS (COVID). These profiles were chosen deliberately to challenge the neural network to classify images with obvious qualitative differences (HPE vs ARDS) and with no obvious differences (NCOVID vs COVID) between their B lines patterns (Figure 1 and online supplemental files 1–3). The COVID class consisted of confirmed cases of COVID-19 via reverse-transcriptase PCR test. The NCOVID class consisted of an assortment of causes: aspiration, community-acquired pneumonia, hospital-acquired pneumonia and viral pneumonias. Examinations were conducted as part of patient encounters in the emergency department, intensive care unit and medical wards across the two hospitals.
Supplementary video
Supplementary video
Supplementary video
Sample images and lung ultrasound characteristics typical of the three lung pathologies that are the subject of our deep learning classifier (videos available in online supplemental files 1–3).
Candidate examinations for inclusion were identified using a sequential search by two critical care physicians, ultrasound experts (RA, TA) from within the finalised clinical reports of our database of LUS cases (figure 2).
Data acquisition, selection and verification workflow.
Videos from our dataset represented a variety of ultrasound systems with phased array probe predominantly used for acquisition. Videos of the costophrenic region (which included solid abdominal organs, diaphragm or other pleural pathologies such as effusions or translobar consolidations) were excluded as (1) these regions did not contribute greatly to alveolar diagnoses, (2) this would introduce heterogeneity into the still image data and (3) a trained clinician can easily distinguish between these pathologies and B lines. Duplicate studies were discarded to avoid overfitting. From each encounter, deidentified mp4 loops of B lines, ranging from 3 to 6 s in length with a frame rate ranging from 30 to 60/s (depending on the ultrasound system), were extracted. As COVID was the newest class available to our database, its comparably smaller number of encounters governed the number of encounters we extracted from HPE and NCOVID. A balanced volume of data for each class of image is important to avoid model overtraining on a single image class and/or overfitting.
Data preprocessing
The images used to train the model were all frames from the extracted LUS clips. Hereafter, a clip refers to an LUS video that consists of several frames. An encounter is considered to be a set of one or more clips that were acquired during the same LUS examination.
Preprocessing of each frame consisted of a conversion to greyscale followed by a script written by one of our teams (JH) to scrub the image of extraneous information (index marks, logos and manufacturer-specific user interface). See online supplemental appendix 1 for full details.
Supplemental material
Data augmentation techniques were applied to images to each batch of training data during training experiments to combat overfitting. Augmentation transformations included random zooming in/out by ≤10%, horizontal flipping, horizontal stretching/contracting by ≤20%, vertical stretching/contracting (≤5%) and bidirectional rotation by ≤10°.
Model architecture and training
In choosing an optimal architecture for our model, we investigated training from scratch on custom implementation of feedforward convolutional neural networks (CNNs), residual CNNs as well as transfer learning methods.17 Ultimately, Xception architecture18 achieved the highest performance among the custom and seven common architectures evaluated.
Individual preprocessed images were fed into the network as a tensor with dimensions of 600×600×3. Although the images were originally greyscale, they were converted to RGB representation to ensure that the model input shape was compatible with the pretrained weights. The output tensor of the final convolutional layer of the Xception model was subject to 2D global average pooling, resulting in a one-dimensional tensor. Dropout at a rate of 0.6 was applied to introduce heavy regularisation to the model and provided a noticeable reduction in overfitting. The final layer was a three-node fully connected layer with softmax activation. The output of the model represents the probabilities that the model assigned to each of the three classes, all summing to 1.0. The argmax of this probability distribution was considered to be the model’s decision. To further combat overfitting, early stopping was applied by halting training if the loss on the validation set did not decrease over the most recent 15 epochs.19
For additional details on model selection, training, coding practice, our GitHub repository and hardware used in this project, see online supplemental appendix 1.
Validation strategy
A modification of the holdout validation method was used to ensure that the model selection process was independent of the model validation. Our holdout approach began with an initial split that randomly partitioned all encounters into a training set and two test sets (henceforth referred to as test-1 set and test-2 set). The distribution of encounters and frames after this split is shown in table 1. Test-1 was used to evaluate all of the candidate models so that a final model architecture and set of hyperparameters could be selected and was then incorporated into the training set for the final validation. Test-2 was held back during model selection and was used to evaluate the model for the final validation phase. Validation sets for both experiments were derived as a subset of the encounters in the training set. A full account of validation methods can be found in the online supplemental appendix 1. It must be emphasised that by splitting sets by encounters, frames from the same encounter (and therefore the same clip) would only be found in the same set during any given experiment.
Distribution of clips and images assigned to each dataset
Measuring model performance
The final model performance was determined by its results on our hold-back, independent dataset (test-2). The results were analysed both at the individual frame level and at the encounter level. The latter was achieved through averaging the classifier’s predicted probabilities across all images from within that encounter. We assessed the model’s performance by calculating the area under the receiver operating characteristic curve (AUC), analysing a confusion matrix and calculating metrics derived from the confusion matrix.
Human benchmarking
Benchmarking human performance for comparison to our model was undertaken using a survey featuring a series of 25 LUS clips from 25 different patients, varying from normal lung (four clips) to different LUS findings (six hydrostatic pulmonary edema HPE, 7 non-COVID pneumonia and 8 COVID pneumonia). All clips were sourced and labelled with agreement from three ultrasound fellowship trained physicians (MW, TA and RA; see online supplemental appendix 1 for complete survey). As to provide every advantage to the human interpretation exercise, survey clips—chosen from among the global study data—that particularly exemplified the characteristics described in figure 1 were chosen for the survey. The survey was distributed to 100 LUS-trained acute care physicians from across Canada. Respondents were asked to identify the findings in a series of LUS loops according to the presence B lines versus normal lung (A line pattern), the characteristics of the pleural line (smooth or irregular) as well as the cause of the LUS findings. Responses were compared with the true, expert-defined labels consistent with our data curation process described above. The four clips of normal lung used were eventually discarded from analysis since the data used for modelling our algorithm did not include normal lungs. Any normal diagnoses (37 of 1281 diagnoses) for the remaining clips were replaced with uniformly randomly generated diagnoses for the remaining causes.
Explainability
We used the Gradient-weighted Class Activation Mapping (Grad-CAM) method to visually explain the model’s predictions.20 Grad-CAM involves visualising the gradients of the prediction of a particular image with respect to the activations of the final convolutional layer of the CNN. A heatmap is produced that is upsampled to the original image dimensions and overlaid onto the original image. The resultant heatmap highlights the areas of the input image that were most contributory to the model’s classification decision.
Data statement
The GitHub link to the code used to generate the DL model and the full survey data results can be found in our online supplemental appendix 1.
Patient and public involvement
Patients or the public were not involved in the design, conduct, reporting or dissemination plans of this work.
Results
Ultrasound data
The data extraction process resulted in 84 cases of COVID (185 loops, average 2.23 loops/case) which, as part of our effort to balance the groups for unbiased training, led to 78 of NCOVID (236 loops, average 2.91 loops/case) and 81 (191 loops, average 2.42 loops/case) of HPE. All data originated from point-of-care, battery-operated machines primarily using a phased array transducer, abdominal imaging preset at an imaging frequency between 2 and 5 megahertz (MHz). Images had similar imaging depths and, due to manufacturer standards, the focal zone was automatically set. With those machines allowing for manual focal zone control, the focus was directed at the pleural line. A variety of different clinician-sonographers obtained the data, as part of their clinical work. Further characteristics of the data, including patient demographics, are summarised in table 2.
Data profile for the three groups of lung ultrasound images used to train and test our model
Human benchmarking
The benchmarking survey was completed by 61 physicians with a median of 3–5 years of ultrasound experience, the majority of whom had done at least a full, dedicated month of ultrasound training (80.3%) and who described their comfort with LUS use as ‘very comfortable’ (72.1%). See online supplemental appendix 1 for a full summary of survey data.
The results of this survey highlight that the physicians were adept at distinguishing the HPE class of B lines from COVID and NCOVID causes of B lines. For the COVID and NCOVID cases, however, significant variation and uncertainty was demonstrated (see table 3 and the ‘Comparing human and neural networks’ section).
Confusion matrices for the physicians (survey responses from 61 physicians classifying lung ultrasound clips into their respective causes, numbers in parenthesis reflect classifications from the aggregated approach used to calculate area under the receiver operating characteristic curve), model performance on the test-2 holdback set at the frame and the encounter level
Model performance on holdback data
The model’s predictions were evaluated at both the image and the encounter levels. The prediction for an image is the probability vector  obtained from the output of the softmax final layer, and the predicted class was taken to be
obtained from the output of the softmax final layer, and the predicted class was taken to be  . Prediction for an encounter was considered to be
. Prediction for an encounter was considered to be  , where
, where  is the average predicted probability for class c over the predictions for all images within that encounter. Encounter-level predictions were computed and presented to (1) replicate the method through which real time interpretation (by clinician or machine) occurs with ultrasound by aggregating images within one or more clips to form an interpretation and (2) closely simulate a physician’s classification procedure, since the physicians who participated in our benchmarking survey were given entire clips to classify. Three models fit with our chosen architecture and set of hyperparameters were evaluated on test-1, achieving mean AUCs on the encounter level of 0.966 (COVID), 0.815 (NCOVID) and 0.902 (HPE). The model’s ultimate ability was to be determined on the 10.1% of our images that constituted the holdback data (test-2) data. On these independent data, the model demonstrated a strong ability to distinguish between the three relevant causes of B lines with AUCs at the encounter level of 1.0 (COVID), 0.934 (NCOVID) and 1.0 (HPE), producing an overall AUC of 0.978 for the classifier. Confusion matrices on the test-2 set at the frame and encounter levels (table 3) show strong diagonals that form the basis of these results and the performance metrics seen in table 4.
is the average predicted probability for class c over the predictions for all images within that encounter. Encounter-level predictions were computed and presented to (1) replicate the method through which real time interpretation (by clinician or machine) occurs with ultrasound by aggregating images within one or more clips to form an interpretation and (2) closely simulate a physician’s classification procedure, since the physicians who participated in our benchmarking survey were given entire clips to classify. Three models fit with our chosen architecture and set of hyperparameters were evaluated on test-1, achieving mean AUCs on the encounter level of 0.966 (COVID), 0.815 (NCOVID) and 0.902 (HPE). The model’s ultimate ability was to be determined on the 10.1% of our images that constituted the holdback data (test-2) data. On these independent data, the model demonstrated a strong ability to distinguish between the three relevant causes of B lines with AUCs at the encounter level of 1.0 (COVID), 0.934 (NCOVID) and 1.0 (HPE), producing an overall AUC of 0.978 for the classifier. Confusion matrices on the test-2 set at the frame and encounter levels (table 3) show strong diagonals that form the basis of these results and the performance metrics seen in table 4.
Classification performance metrics calculated from the model’s predictions and ground truth from the test-2 set
Comparing human and neural network results
We compared the physician-predicted results to our model’s test-2 results. Since AUC measures a classifier’s ability to rank observations, the raw survey data (in the form of classifications, not probabilities) were processed to permit an AUC computation by considering physician-predicted probability of a LUS belonging to a specific class as the proportion of physicians that assigned the LUS to that class. The AUCs for the physicians, at face value, were 0.697 (COVID), 0.704 (NCOVID) and 0.967 (HPE), leading to an overall AUC of 0.789 (as compared with 0.978 for our model). A comparison of the human and model AUCs is graphically displayed in figure 3. We took note of the AUC of approximately 0.7 for the physicians when the positive class is COVID or NCOVID, as distinguishing between these classes is not known to be possible by humans. In examining the raw confusion matrix data (table 3), this suggests near random classification (which corresponds to an AUC of 0.5) between these two classes, see online supplemental appendix 1 for a complete explanation. Given the important implications of the performance gap observed, we employed an additional step of statistical validation for our findings through a Monte Carlo simulation (MCS, see online supplemental appendix 1 for full details) of human performance, based on our survey results, across 1 million exposures to our test-2 data.21 After simulating this performance 1 million times, the MCS yielded an average AUC of 0.840 across all three classes, with very few cases matching or exceeding the performance of the CNN. Thus, we can conclude that our model exceeds human performance, and in particular, that the model can distinguish between COVID and NCOVID (p<0.01).
Receiver operating characteristic curves across the three classes of images that our human benchmarking (physicians) and our model (convolutional neural network (CNN)) were tasked with interpreting. The model’s performance on the test-2 (holdback) image set is plotted for both individual images and across the entire image set from one encounter. In all image categories, it can be seen that the model interpretation accuracy exceeded that of the human interpretation.
Explainability results
The Grad-CAM explainability algorithm was applied to the output from the model on the holdback data. The results are conveyed by colour on the heatmap, overlaid on the test-2 input images. Blue and red regions correspond to highest and lowest prediction importance, respectively. As the results in figure 4 show, the key activation areas for all classes were centred around the pleura and the pleural line.
Grad-CAM heatmaps corresponding to a selection of our model’s predictions. Blue areas reflect the regions of the image with the highest contribution to the resulting class predicted by the model. In all cases, the immediate area surrounding the pleura appears most activated. COVID, COVID-19 pneumonia; HPE, hydrostatic pulmonary edema; NCOVID, non-COVID-related acute respiratory distress syndrome.
Discussion
In this study, a DL model was successfully trained to distinguish the underlying pathology in similar point-of-care LUS images containing B lines. The model was able to distinguish COVID-19 from other causes of B lines and outperformed ultrasound-trained clinician benchmarks across all categories. Our results, within the context of the limitations outlined below, are the first of their kind to support that digital biomarker profiles may exist within LUS images.
Our model was developed using a dataset of 243 patients (612 video loops/121 381 frames) which is modest by machine learning standards. Owing to the scarcity of labelled LUS data, this data volume does compare favourably to other published LUS work.22–24 Given the implications of successfully classifying LUS images, it was essential for us to protect against overfitting. While many approaches exist to avoid an overfit model, we, in addition to multiple data augmentation techniques, reserved 10% of our data (test-2) as a holdback set, not involved in model fitting or selection. This approach mimics the unbiased, generalisable performance desired of an image classifier and is familiar to other notable DL vision research in medicine.9–11
DL has shown similarly favourable results in recent CXR and CT studies of COVID-19.25 26 Given LUS image creation is fundamentally different (producing artefacts, rather than anatomic images of the lung), it could not be expected that our work with LUS would have yielded such similar results. The value of identifying such accuracy in an LUS model rests in the ability of LUS (unlike CT or CXR) to be delivered by limited personnel, at low cost and in any location.
LUS artefact analysis has existed for several years in some commercially available ultrasound systems and has also been described using various methods in the literature.22 27 28 Automating the detection of canonical findings of LUS, these techniques are convenient and serve to achieve what clinicians may be trained to do with minimal training.29 With attention to COVID-19, LUS has been shown to inform clinical course and outcome,30 creating some further momentum towards broader LUS competence. As our work opens the door toward plausible early, automated COVID identification using LUS, DL techniques to autogenerate clinical severity score for COVID has also recently been described.23 The eventual integration of various DL models into ultrasound hardware seems plausible as a method to achieve real-time, point-of-care diagnosis and prognosis of COVID or other specific respiratory illnesses.
The implications of our work, at the time of writing, are strongly attached to the current challenges and importance of COVID-19 diagnosis. Our results point to a unique, pixel-level signature within the COVID-19 image. Although the exact mechanism of distinction is unknown, the heatmap results suggest that subvisible variations in the pleural line itself is most active in driving the model’s performance. The value of Grad-CAM heatmaps in explaining DL work on LUS has recently been highlighted by other experts in the field.31 The precise taxonomical implications of our findings, whether they are driven by COVID-19, coronaviruses or viruses a whole, will require additional research.
Our study has some important limitations. The first relates to the opaqueness that is implicit to deep neural networks. Despite using Grad-CAM, the decisions by the trained model are not outwardly justified and we are unable to critique its methods and must trust its predictions. Our benchmarking survey did not exactly replicate the questions posed to our neural network which made our statistical analysis more complex than it might have needed to be otherwise. The other limitations of our study are related to the data. Although our model performance signal was strong, the addition of further training data can only aid with generalisability of the model. Further, as our data were not prospectively acquired, we lacked the ability to standardise its characteristics, resulting in heterogeneous imaging properties. The use of non-standard imaging depths and imaging frequencies produces sufficient heterogeneity which, combined with the inability to precisely audit the learning points of the CNN, does introduce the risk that the basis of our results could be driven by these variations rather than the variations in B line artefacts. Given that heterogeneous data can introduce such strong bias to model performance, a proposed pathway for standardised LUS image acquisition may serve as a roadmap for future DL work.32 By setting standards, including imaging sets, shared international database, standard LUS interpretation scores and the use of linear or convex ultrasound transducers (rather than phased array, which was used predominantly in our work and is common in North America) will minimise bias and enrich the results of future scholarship in DL and LUS. Lastly, our data were all from hospitalised patients and our results may not generalise to those who are less ill.
Conclusions
With strong performance in distinguishing LUS images of COVID-19 from mimicking pathologies, a trained neural network exceeded human interpretation ability and raises the possibility of disease-specific, subvisible features contained within LUS images. To confirm these findings, research using homogeneous, well-labelled, multicentre data is indicated.
Acknowledgments
The authors would like to acknowledge the computational and technical support from CENGN (Canada’s Centre of Excellence in Next Generation Networks), Mr Matt Ross from the City of London, Mrs Kristine Van Arsen from the Division of Emergency Medicine and the clinician-sonographers at London Health Sciences Centre who faithfully record and annotate their lung ultrasound studies.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Twitter @arntfield
Contributors All authors were involved in the authorship of the manuscript, figures and tables. Overall project design and oversight (RA, BV), data management (TA, DW, RA, JH, MW), survey creation and distribution (MW), model training (BV, DW, NP), statistical analysis (NP), figure generation (JH, NP, BV) and literature search (RC, RA).
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests None declared.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available in a public, open access repository. Data are available upon reasonable request. The code for the neural network is available at the below URL. This code may be used to further this line of work. https://github.com/bvanberl/covid-us-ml Deidentified ultrasound images are not contained in an open repository due to the volume of data. Inquiries about this data may be directed to the corresponding author of this paper.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.