Article Text
Abstract
Introduction Our aim is to develop a novel approach to hyperkinetic movement disorder classification, that combines clinical information, electromyography, accelerometry and video in a computer-aided classification tool. We see this as the next step towards rapid and accurate phenotype classification, the cornerstone of both the diagnostic and treatment process.
Methods and analysis The Next Move in Movement Disorders (NEMO) study is a cross-sectional study at Expertise Centre Movement Disorders Groningen, University Medical Centre Groningen. It comprises patients with single and mixed phenotype movement disorders. Single phenotype groups will first include dystonia, myoclonus and tremor, and then chorea, tics, ataxia and spasticity. Mixed phenotypes are myoclonus-dystonia, dystonic tremor, myoclonus ataxia and jerky/tremulous functional movement disorders. Groups will contain 20 patients, or 40 healthy participants. The gold standard for inclusion consists of interobserver agreement on the phenotype among three independent clinical experts. Electromyography, accelerometry and three-dimensional video data will be recorded during performance of a set of movement tasks, chosen by a team of specialists to elicit movement disorders. These data will serve as input for the machine learning algorithm. Labels for supervised learning are provided by the expert-based classification, allowing the algorithm to learn to predict what the output label should be when given new input data. Methods using manually engineered features based on existing clinical knowledge will be used, as well as deep learning methods which can detect relevant and possibly new features. Finally, we will employ visual analytics to visualise how the classification algorithm arrives at its decision.
Ethics and dissemination Ethical approval has been obtained from the relevant local ethics committee. The NEMO study is designed to pioneer the application of machine learning of movement disorders. We expect to publish articles in multiple related fields of research and patients will be informed of important results via patient associations and press releases.
- neurology
- neurophysiology
- adult neurology
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Strengths and limitations of this study
This cross-sectional study addresses a real need in classification of hyperkinetic movement disorders and may revolutionise our clinical approach.
Representative patients from the entire hyperkinetic movement disorders spectrum will participate, including myoclonus, dystonia, tremor, chorea and tics, as well as ataxia, spasticity and functional movement disorders.
An extensive data set will be collected and used for analysis: clinical information and data from electromyography, accelerometry and three-dimensional video.
The project is a collaboration between movement disorders specialists at an acknowledged expertise centre and computer scientists in the corporate and academic world.
Calculation of the needed sample size is hampered by the absence of previous studies in this exact field.
Introduction
The hyperkinetic movement disorders myoclonus, dystonia, tremor, chorea and tics are all characterised by excessive, involuntary movements (table 1).1 In a patient presenting with such a disorder, it is crucial to classify the movement disorder phenotype promptly and accurately, because the phenotype classification will guide clinical decisions on diagnostic testing and (symptomatic) treatment.1 Current clinical algorithms on hyperkinetic movement disorder diagnosis, whether aimed at dystonia,2 myoclonus3 or tremor,4 all start with the same step: make sure to establish the main movement disorder phenotype in the patient. The subsequent diagnostic steps consist of additional tests such as laboratory, imaging and genetic tests, which can greatly aid the patient’s aetiological diagnosis. The same holds for treatment: decisions on which medication to prescribe or which target to choose for deep brain stimulation are directed by the main clinical movement disorder phenotype.
Hyperkinetic movement disorders and related disorders
Given the importance of clinical phenotype recognition, it is unfortunate that correct classification is difficult in many patients. While each phenotype of movement disorder has its own clinical presentation, complex and mixed forms occur. Moreover, the big five hyperkinetic phenotypes (myoclonus, dystonia, tremor, chorea and tics) may share overlapping clinical features with ataxia, spasticity and functional movement disorders. Presently, the classification of involuntary movements is based on clinical definitions and therefore on expert opinion. However, research demonstrated large interobserver and intraobserver variability of clinical classification.5–8 This is a major problem that impairs correct phenotyping of patients which subsequently delays aetiological diagnosis, deliverance of tailored treatment and evaluation of treatment effects.
To improve the classification of hyperkinetic movement disorders we set-up the current study, Next Move in Movement Disorders (NEMO). Our aim is to combine electromyography (EMG), motion sensors and three-dimensional (3D) video with machine learning to develop a computer-aided classification tool for hyperkinetic movement disorders, which will help healthcare professionals establish the movement disorders phenotype. The algorithm of this computer-aided classification tool will be trained with the input data from patients who have a hyperkinetic movement disorders phenotype on which a panel of blinded independent experts agree.
Currently, EMG and accelerometry are applied in clinical practice to support the clinical classification of movement disorders, particularly myoclonus, tremor and dystonia. This is done by assessment of features such as muscle activation pattern, burst duration and frequency. While not all applications are supported by extensive evidence, there is a long tradition and ample experience with these methods.9 Despite the frequently incorporated clinical application of video to discuss cases among panels of experts, the use of video to automatically detect, quantify and classify involuntary movements is still in its infancy. Some advances have recently been made in using video for assessment of tremor frequency in a research setting.10 While these first steps are encouraging, application in other movement disorders is still lacking. Moreover, until now, investigations of machine learning in movement disorders have mainly focused on quantification of the hyperkinetic movements11 and optimisation of adaptive deep brain stimulation.12 Efforts to classify different types of hyperkinetic movement disorders have not yet been made, to our knowledge.
In the NEMO project, we combine EMG, accelerometry and video recordings to develop an innovative new approach to movement disorder classification. We see the development of a computer-aided classification tool as the next step towards swift and accurate phenotype classification of hyperkinetic movement disorders, the cornerstone of both the diagnostic and treatment process.
Methods
Study population
The NEMO study protocol comprises patients with both single and mixed phenotype movement disorders. Single phenotype groups will primarily include dystonia, myoclonus and tremor, and in the future chorea, tics, ataxia and spasticity will be added. Although spasticity is not typically classified as a movement disorder, this group is added for distinction from dystonia, as these two neurological conditions are sometimes confused with one another.6 Similarly, ataxia is not usually listed as a hyperkinetic movement disorder, but signs of ataxia can overlap with symptoms of hyperkinetic movement disorders, making it a relevant phenotype to include.13 Mixed phenotype groups will consist of myoclonus-dystonia, dystonic tremor, myoclonus ataxia and jerky/tremulous functional movement disorders. All patient groups will consist of 20 participants. In addition, a group of 40 healthy participants will be included. Patients will be carefully selected based on all clinical information (MRK, AMMvdS and MAJT): after participating in the experiment, their data will be reviewed by a panel of experts to assess their phenotype and whether they meet the gold standard for inclusion.
Participants will only be eligible for inclusion if they are at least 16 years old. Exclusion criteria consist of comorbid conditions that lead to impaired upper extremity function, and a silver allergy or implanted pacemaker due to incompatibility with the EMG device. Moreover, healthy participants cannot be first-degree relatives of patients with hyperkinetic movement disorders.
Recruitment
Patients will be selected from the University Medical Center Groningen (UMCG) hyperkinetic movement disorders database and recruited at the UMCG outpatient clinic. If patient inclusion using these two sources does not lead to the targeted number of patients, we will recruit at other hospitals and via patient associations. Healthy participants will also be recruited at the UMCG. Participants will receive written information about the study and will have the opportunity to ask the investigators questions beforehand. Prospective participants can also call on an independent neurologist, who is not involved in the NEMO study, if they have any remaining questions. All participants will give written informed consent before enrolment.
Study setting
Movement Disorders Groningen, based at the UMCG, is uniquely positioned for the conduction of the NEMO study, as we treat patients with a broad range of hyperkinetic movement disorders. Movement Disorders Groningen is acknowledged by the Dutch Federation of University Medical Centres (NFU) as an official centre of expertise, and it is part of the European Reference Network for rare diseases. Data analysis will be performed in collaboration with ZiuZ Visual Intelligence BV (IG and JC), and the departments of computing science at the University of Utrecht (ACT) and the University of Groningen.
Experimental set-up
Clinical information
Clinical information such as age at onset, family history, disease progression, medication use, symptom influencing factors and psychological reports will be acquired from the participants during their study visit. Moreover, the influence of their movement disorder on activities of daily life will be assessed using the Fahn-Tolosa-Marin Essential Tremor Rating Scale part C,14 with additional questions that originate from the Burke-Fahn-Marsden Dystonia Rating Scale,15 SARA Ataxia Scale,16 Unified Myoclonus Rating Scale,17 Shapiro Tourette’s Syndrome Severity Scale18 and Abnormal Involuntary Movement Scale.19 Additionally, patients will be asked to indicate the perceived severity of their movement disorder on a Visual Analogue Scale.
Movement tasks
Data will be collected while participants are performing several movement tasks. These tasks are selected from the Fahn-Tolosa-Marin Essential Tremor Rating Scale,14 Burke-Fahn Marsden Dystonia Rating Scale,15 SARA Ataxia Scale,16 Unified Myoclonus Rating Scale,17 Shapiro Tourette’s Syndrome Severity Scale18 and Abnormal Involuntary Movement Scale.19 All of these are validated scales, used both in clinical practice and in experimental settings to assess the severity of specific movement disorders. As such, they contain tasks that are designed to elicit the movement disorder in a patient, enabling assessment of severity. Selection of tasks was discussed in depth with seven movement disorders, neuropaediatric and neurorehabilitation specialists with particular experience in the field of dystonia, myoclonus, tremor, chorea, tics, ataxia or spasticity to ensure comprehensive coverage of all disorders. Table 2 lists the set of tasks to be included. A trained investigator will instruct the participant before the recording of each task. If the task is not performed as instructed or the data collection was suboptimal, the patient is asked to repeat the task. Version control of tasks is used to ensure that the correct version of the task is included in the data analysis.
Included tasks
Data collection
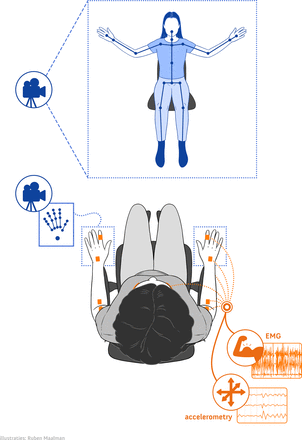
Data from EMG, motion sensors and 3D video will be collected simultaneously using custom made software (Visual Studio, Microsoft, USA). Surface EMG and motion sensor data will be recorded using the Trigno system (Delsys, Massachusetts, USA). Data collection frequencies of the EMG and the motion sensors are 2000 Hz and 150 Hz, respectively. The Trigno Avanti sensors measure muscle activity, 3D accelerometry, 3D gyroscope and 3D magnetometry in each single wireless device. Ten Trigno Avanti sensors will be placed bilaterally on the participant’s arms and neck, aiming for the musculus (m.) biceps, m. triceps, m. extensor carpi radialis, m. flexor carpi ulnaris and on both index fingers for accelerometry purposes only. Additionally, the Trigno Mini sensors measure muscle activity, using the sensor head and 3D accelerometry, using the sensor base, in each single wireless device. The six Trigno Mini sensor heads, registering EMG data, will be placed bilaterally aiming for the m. abductor pollicis, m. abductor digiti quinti and m. sternocleidomastoideus. The six Trigno Mini sensor bases, registering accelerometry, will be placed bilaterally at the inside of the forearm near the wrist, the back of the hand and the jaw. Figure 1 illustrates the placement of the EMG and motion sensors.
Depiction of sensor placement.
3D video data will be recorded using two types of 3D video cameras. A Realsense D435 camera (frame rate 30 fps; Intel, California, USA) will be used for whole body tracking; two Leap Motion cameras (frame rate 120 fps; Leap Motion, California, USA) will be employed to track the hands specifically. For the Realsense D435 camera, the Nuitrack API (3DiVi, California, USA) is used to extract joints from the depth frames. Aside from the depth frames, the Realsense D435 camera also collects two-dimensional (2D) colour frames at 30 fps. The Leap Motion camera uses an internal algorithm to extract joints from the depth frames. Figure 2 depicts which body parts are tracked by the cameras.
Depiction of the study set-up and data collection. EMG, electromyography.
The EMG, motion sensors and 3D video data all have different sampling frequencies or frame rates. To enable synchronisation of modalities, a time stamp of each data sample point is saved during data acquisition.
Sample size
For the current study, artificial intelligence experts expect that 20 subjects per patient group will be sufficient to develop proof-of-principle for a computer-aided diagnosis tool, because of the extensive amount of data that is collected per participant. Calculation of the needed sample size is hampered by the absence of previous studies in this exact field. While machine learning with, for example, MRI20 and EEG21 data has previously been used to classify other diseases, both these data and disorders differ from the proposed study and are therefore not suitable to perform power analyses.
Data management
Study data will be handled confidentially and in accordance with the European personal data protection regulation (General Data Protection Regulation). A unique identification code will be assigned to each participant. All raw de-identified EMG, motion sensor and 3D video data will be stored at the research drive hosted and provided by the UMCG. Preprocessed data will be stored and analysed in the virtual research workspace, a secure virtualised desktop environment provided and hosted by the University of Groningen. De-identified clinical data are collected and managed using REDCap electronic data capture tools hosted at the UMCG.22 23
Data analysis
Gold standard
The ‘gold standard’ for phenotype classification is defined as interobserver agreement among three experienced (inter)national clinical experts who will each assess the phenotype of a patient independently. Their evaluation will be based on the video recordings (RGB, 2D) of the participants performing all the different tasks described in table 2 during the experiment. Moreover, a sound fragment is recorded of the patient’s speech for the experts’ assessment. They will also be presented with information from the history and ancillary tests, to mimic the typical outpatient clinical situation as best as we can. The experts will be blind to the phenotype classification made by the investigators. All video data for the experts can be reviewed using the virtual research workspace and the relevant clinical information can be assessed in REDCap. Interobserver agreement among the clinical experts will be calculated using Fleiss’ kappa (K).24 We consider a Fleiss’ K value above 0.80 (interpreted as excellent agreement) across all groups as sufficient. Moreover, for each individual participant, a minimum of two out of three experts should be in agreement. If a participant does not meet this requirement, their data will be excluded from further analysis.
Machine learning analyses
To develop the computer-aided classification tool, several machine learning techniques will be used. The clinical parameters, EMG, motion sensors and 3D video data will serve as input for the machine learning algorithm. The expert-based phenotype classification will serve as patient labels for supervised learning. In supervised machine learning, an algorithm is trained with multiple examples with a known output label (the expert-based phenotype classification), learning how to process the input data (clinical, EMG, motion sensor and video data) to reproduce the output label. The fully trained algorithm can then be given new input data, and is able to predict what the output label should be, making these types of algorithms well suited for classification.25 Examples of machine learning techniques that will be investigated include deep learning methods such as long short-term memory networks26 and methods based on manually engineered features such as random forest classifiers27 28 and learning vector quantisation29 30 using features from clinical neurophysiology research and practice (eg, frequency analysis or intermuscular coherence). From the above classes, deep learning methods have the advantage of computational scalability and the ability to automatically extract the relevant features for classification. In contrast, methods using manually engineered features allow one to control which aspects of the data are used by the classifier to reach its decisions and thus can be trained with fewer data to arrive at high accuracies. The information about how machine learning algorithms arrive at their output labels is often limited, which can mask confounding factors or simply diminish trust in the produced outputs. To address this problem, we will employ visual analytics to visualise how the classification algorithm arrives at its decision, and in particular which data aspects it uses to reach that decision.
Ethics and dissemination
The NEMO study protocol has been approved by the Medical Ethical Committee (METc 2018–444) of the UMCG and will be conducted according to the ethical standards of the Helsinki Declaration.
This study is designed to pioneer the application of machine learning in the classification of movement disorders, and allows for comprehensive data collection of clinical information, EMG and motion sensor data and 3D video. Because of its scope, we expect to publish multiple articles in the fields of clinical neurology, particularly movement disorders, clinical neurophysiology, artificial intelligence and visual analytics. Moreover, patients will be informed of important study results via the different patient associations, press releases, the website www.movementdisordersgroningen.com and at the Movement Disorders Groningen biannual Patient Day.
Patient and public involvement
Members of the Movement Disorders Groningen Patient Council are updated regularly about the progress of the NEMO study and advise on online patient information. The help of several patient associations is enlisted for the recruitment of participants. Dissemination to patients and the general public is described above.
Discussion
The NEMO study seeks to develop a computer-aided classification tool for hyperkinetic movement disorders. Such a tool would aid doctors in establishing the movement disorders phenotype. As adequate phenotype classification is the foundation of clinical decision-making and treatment of hyperkinetic movement disorders, improving this process would ultimately result in swifter and more accurate diagnoses and treatment for patients. To this end, we will investigate patients with myoclonus, dystonia, tremor, chorea, tics and functional jerky/tremulous movement disorders, as well as the related disorders ataxia and spasticity.
Our study has several strengths and some possible limitations. A major strength of this study is that it is conducted at a leading centre of expertise for movement disorders, Movement Disorders Groningen at the UMCG, The Netherlands. Experts in the field of all hyperkinetic movement disorders are present for collaboration and patient recruitment. For example, several experts assisted in selection of the appropriate tasks that patients will perform during the experiment, ensuring proper investigation of all relevant movement disorders. Also, the centre has a large patient database, which is a substantial advantage as some movement disorders are rare. Moreover, we have good rapport with our centre’s patient council, who can advise on recruitment and dissemination of results. Disease rarity might still limit our speed of inclusion, a potential limitation, but given the advantages linked to our centre of expertise as well as the collaborations that exist with other centres, we are confident we will include enough patients in each group.
A second significant strength is the collection and analysis of extensive data: both clinical information and data from EMG, motion sensors and 3D video. In the analysis of these data, we can make use of the expertise of several members in our team. For instance, we have ample experience with the analysis of EMG and motion sensor data in movement disorders patients at the department of clinical neurophysiology.31–34 Similarly, ZiuZ Visual Intelligence BV, a major collaborator in this project, has a proven track record of developing and deploying systems that collect and analyse visual data in different sectors and thus the required expertise for designing and implementing the computer-aided diagnosis tool as well as the data acquisition software for NEMO. In addition, academic input on the topic of machine learning and visual analytics is available from the University of Utrecht35–38 and the University of Groningen.39–42
Significance
The aim of this study is to move the classification of movement disorder phenotypes forward, to improve the diagnostic process and optimise treatment in patients with hyperkinetic movement disorders. This could be achieved by the development of an all-encompassing computer-aided diagnostic tool for all hyperkinetic movement disorder phenotypes, which is the ultimate goal. Such a tool would be a game changer, helping neurologists to establish the phenotype in their patients with more certainty, so they can swiftly move on to other investigations into aetiology and to start the correct treatment.
In the process, it is likely that several new diagnostic biomarkers for hyperkinetic movement disorders will be discovered. These biomarkers can have their bearing on the current clinical practice quite quickly, particularly if they are in the fields of ECG or accelerometry because these techniques are already widely applied in movement disorders diagnostics.
Additionally, the tool that is being developed in the current study could be used to monitor disease severity and progression. This is of relevance for the monitoring of patients during treatment, for instance before and after deep brain stimulation. Another application could be patient assessment in clinical trials.
Study status
Data collection is in progress.
Ethics statements
Patient consent for publication
References
Footnotes
Twitter @maddyvdstouwe
Contributors Conception of the study: MAJdKT and ACT. Design of the experiment: AMMvdS, IT, IG, JC, RG, SvdV, MRK, ACT and MAJdKT. Writing of the article: AMMvdS and IT. Review and critique: IG, JC, RG, JD, SvdV, MRK, ACT and MAJdKT.
Funding This work was supported by the European Fund for Regional Development of the European Union in collaboration with the province of Fryslan (grant number 01492947), and a ZonMW TOP Grant (grant number 91218013).
Competing interests Authors IG, JC and MRK are employed by ZiuZ Visual Intelligence BV, a company with an interest in the development of software tools to aid medical care. Presently, there is no commercial stake in this project.
Patient and public involvement Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review Not commissioned; peer reviewed for ethical and funding approval prior to submission.